Difference between revisions of "Part:BBa B0030"
| Line 12: | Line 12: | ||
== Modelling == | == Modelling == | ||
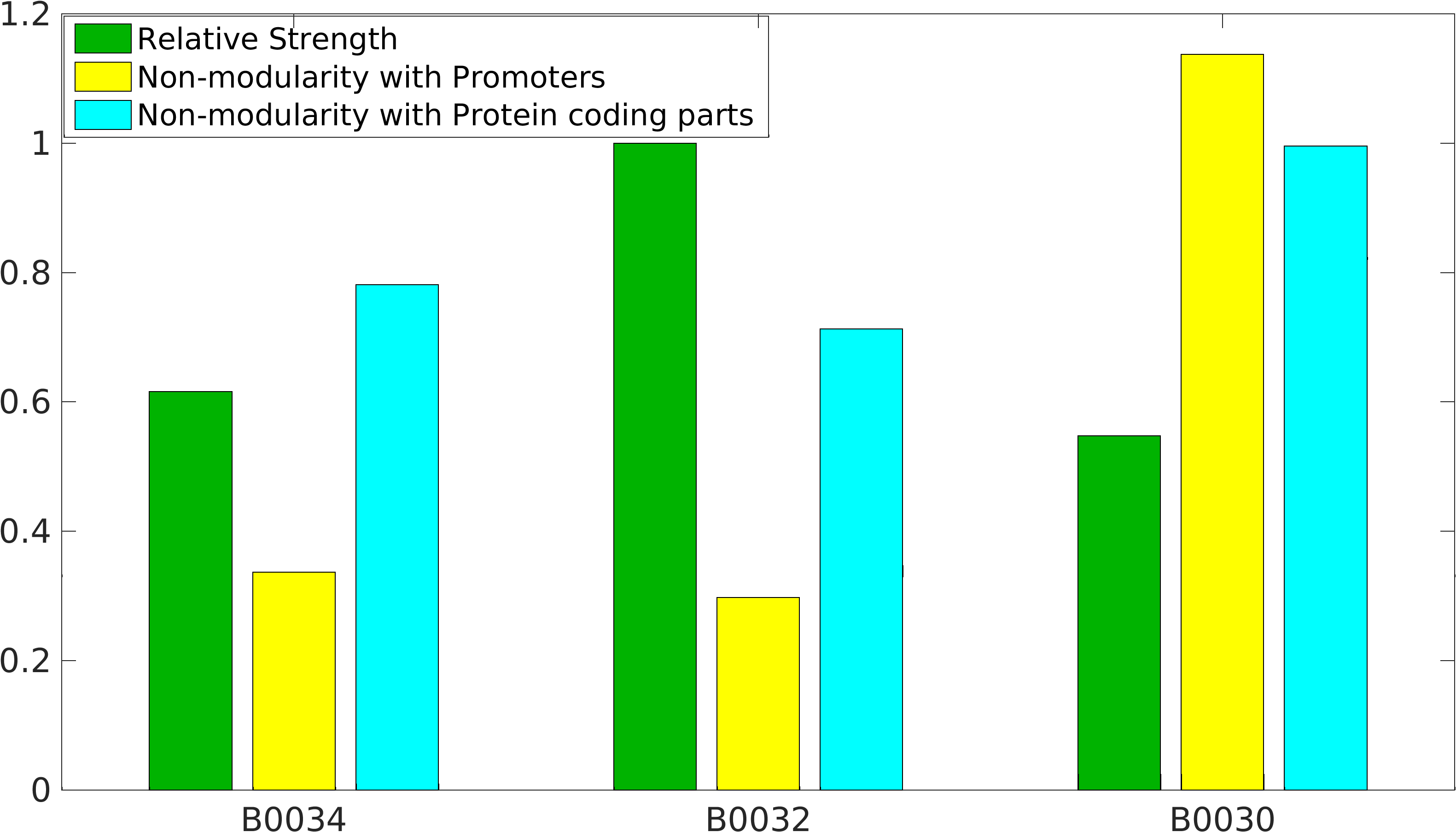
| − | + | Global non-modularity towards promoters & protein coding parts and relative strength was estimated for RBSs B0030, B0032, B0034 in our [http://2016.igem.org/Team:IIT-Madras/Model#Modularity_of_RBS_parts | modelling] | |
== Experimetnation == | == Experimetnation == | ||
Note: not compatible with R0053 promoter due to likely transcript secondary structure This combination yielded very low gfp expression (see BBa_I7108). [jcb 8/3/05]. | Note: not compatible with R0053 promoter due to likely transcript secondary structure This combination yielded very low gfp expression (see BBa_I7108). [jcb 8/3/05]. | ||
Revision as of 21:30, 16 October 2016
RBS.1 (strong) -- modified from R. Weiss
Strong RBS based on Ron Weiss thesis. Strength is considered relative to BBa_B0031, BBa_B0032, BBa_B0033 and BBa_B0034.
Usage and Biology
IIT Madras 2016's Characterization
Modelling
Global non-modularity towards promoters & protein coding parts and relative strength was estimated for RBSs B0030, B0032, B0034 in our [http://2016.igem.org/Team:IIT-Madras/Model#Modularity_of_RBS_parts | modelling]
Experimetnation
Note: not compatible with R0053 promoter due to likely transcript secondary structure This combination yielded very low gfp expression (see BBa_I7108). [jcb 8/3/05].
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Team Warsaw 2010's measurement
RBS strength (relative to B0034): 91,84%Functional Parameters
| biology | -NA- |
| efficiency | 0.6 |