Difference between revisions of "Part:BBa K1618033"
LedaCoelewij (Talk | contribs) |
LedaCoelewij (Talk | contribs) |
||
| Line 5: | Line 5: | ||
| − | The construct was developed using GoldenGate Cloning. These parts were tested by cloning into a binary plasmid vector backbone prior to delivery to plants leaves using Agrobacterium tumefaciens mediated transfection. This was then converted to a standard BioBrick for shipping to the registry by cloning into the Mo-Flipper Plasmid [https://parts.igem.org/Part:BBa_K1467400 BBa_K1467400 ] (complete transcriptional unit flipper) made by the NRP-UEA 2014 team. | + | The construct was developed using GoldenGate Cloning. These parts were tested by cloning into a binary plasmid vector backbone prior to delivery to plants leaves using <i>Agrobacterium tumefaciens</i> mediated transfection. This was then converted to a standard BioBrick for shipping to the registry by cloning into the Mo-Flipper Plasmid [https://parts.igem.org/Part:BBa_K1467400 BBa_K1467400 ] (complete transcriptional unit flipper) made by the NRP-UEA 2014 team. |
Revision as of 20:38, 17 September 2015
Acyltransferase Candidate 1 Expression Construct
This is an acyltransferase candidate with the aim to acylate starch. The construct is made up of P-35s(K1467101)_TP(K1618037):MAO(K1618001)_T-35s(BBa_K1618037)). The construct should be able to transfer acyl groups to starch.
The construct was developed using GoldenGate Cloning. These parts were tested by cloning into a binary plasmid vector backbone prior to delivery to plants leaves using Agrobacterium tumefaciens mediated transfection. This was then converted to a standard BioBrick for shipping to the registry by cloning into the Mo-Flipper Plasmid BBa_K1467400 (complete transcriptional unit flipper) made by the NRP-UEA 2014 team.
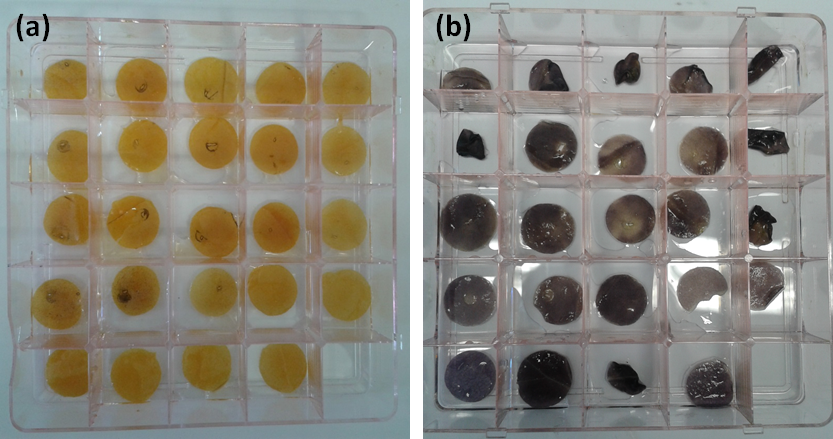
NRP-UEA 2015 originally used this construct with the aim to characterise the starch and determine if acylation had occurred. First, we had to determine if the presence of the enzymes would interfere with starch production. We transformed our constructs into Agrobacterium tumefaciens and then infiltrated it into our plants. We then Iodine stained our leaves to analyse the starch content in the leaves (Figure 1).
Figure 1: Iodine staining of leaves (a) after a 24 hour dark treatment and (b) after an 8 hour light treatment, where yellow stains mean no starch and black stains mean starch present. Dark treatment was performed so the plant would break down its already existing starch storages. This means that all starch found after the light treatment, when the plant is able to build up its stores again, is indicative on the plants ability to produce starch in the presence of our enzymes. The difference between dark and light treatment is evident, as the majority of the dark treatment is stained yellow, while the light treatments were stained completely black. The fact that the controls and each individual enzyme are indistinguishable in the light treatment suggests that none of the enzymes interfere with the formation of starch.
Unfortunately, we were unable to characterise the starch molecules to visualise if acyl groups were added to the starch. However, with a functioning chloroplast transit peptide, we know our constructs reach the chloroplast where starch is produced. The results above also suggest that if acyl groups are being added to the starch, they are not interfering with the starch production.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 392
Illegal BglII site found at 1138
Illegal XhoI site found at 1342
Illegal XhoI site found at 2213 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 2005
- 1000COMPATIBLE WITH RFC[1000]