Difference between revisions of "Part:BBa K143012:Experience"
(→User Reviews) |
Emreilpars (Talk | contribs) |
||
| Line 35: | Line 35: | ||
|}; | |}; | ||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K143064 AddReview 3</partinfo> | ||
| + | |width='60%' valign='top'| | ||
| + | This part was used as a part of the Bee subtilis, iGEM METU 2013 project. We did alignment by using CLUSTAL W between two Pveg promoters; K143012 and K823003. The result is as follows: | ||
| + | |||
| + | http://wiki.metuigem.com/images/1375868_10202188366932242_1731194941_n.jpg | ||
| + | |||
| + | K823003 sequence has 237 bp and K143012 sequence has 97 bp as seen in the figure. We can conclude that K823003 includes K143012. After BLASTing the remaining part of the K823003 which is not same as K143012 there were no results for specific sequences for polimerase binding. This shows K823003 has alike promoter strength as K143012. Only the primers were desinged further from the main promoter region. So for this promoter we can use the promoter strength of the K143012. | ||
| + | |}; | ||
<!-- DON'T DELETE --><partinfo>BBa_K143012 EndReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_K143012 EndReviews</partinfo> | ||
Latest revision as of 02:49, 5 October 2013
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K143012
User Reviews
UNIQ9f13105e652b73c7-partinfo-00000000-QINU
|
Korinna |
We (LMU-Munich 2012) wanted to work with this promoter, but the DNA was not available. So we created a new BioBrick Pveg and characterized it with the reporter lux operon. See the evaluation of the new part: For more Details visit our [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks#Constitutive_promoters_from_B._subtilis webpage] Evaluation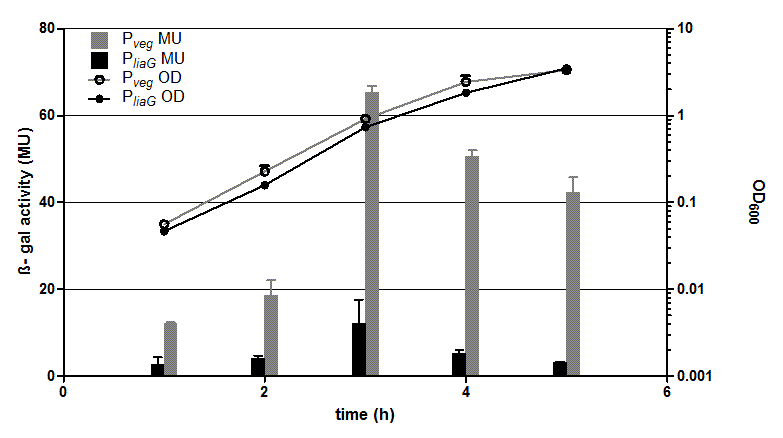 β-galactosidase assay and growth curve of strains carrying the promoters PliaG (black) and Pveg (grey) fused to lacZ. β-galactosidase activity (Miller Units)and the growth curve values are the average of two independant clones with their standard deviation that were measured during the same experiment. Experiment shows representative data which was obtained in the same way from three independent experiments. The two constitutive promoters PliaG and Pveg were evaluated with the reporter lacZ. Promoter activity leads to the expression of the β-galactosidase which directly correlates to the promoter activity. The β-galactosidase assay of the constitutive Bacillus promoters Pveg and PliaG was repeated three times. Data show one representative result. In the beginning of the growth curve both promoters show only low activity. But then it increases to a maximum before it decreases to the begininng level after about seven hours (Data not shown). Summing up the course of activity of both promoters Pveg and PliaG is very similar based on the growth curve. The highest β-galactosidase activity and therefore the highest activity of the promoter Pveg with a maximum of 65 Miller units can be found during the transition from the logarithmic to the stationary phase. This is about five times higher than the acitivity of the promoter PliaG with a maximum activity of about 12 Miller Units. |
|
•••••
Chris |
This part was used as part of a promoter and RBS pair (BBa_K143053) as part of the Biofabricator subtilis Imperial iGEM 2008 project. When incorporated into GFP and RFP production constructs(BBa_K143079 and BBa_K143082), the promoter and RBS pair successfully produced a GFP or RFP output. |
|
•••
|
This part was used as a part of the Bee subtilis, iGEM METU 2013 project. We did alignment by using CLUSTAL W between two Pveg promoters; K143012 and K823003. The result is as follows: http://wiki.metuigem.com/images/1375868_10202188366932242_1731194941_n.jpg K823003 sequence has 237 bp and K143012 sequence has 97 bp as seen in the figure. We can conclude that K823003 includes K143012. After BLASTing the remaining part of the K823003 which is not same as K143012 there were no results for specific sequences for polimerase binding. This shows K823003 has alike promoter strength as K143012. Only the primers were desinged further from the main promoter region. So for this promoter we can use the promoter strength of the K143012. |
