Difference between revisions of "Part:BBa K1022101"
| Line 25: | Line 25: | ||
'''Result:''' | '''Result:''' | ||
| − | |||
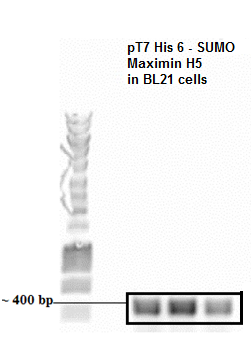
[[Image:Maximin H5.png|300px|center]] | [[Image:Maximin H5.png|300px|center]] | ||
:::::::::::::: ''' Colony PCR SUMO Peptide ''' | :::::::::::::: ''' Colony PCR SUMO Peptide ''' | ||
Revision as of 20:11, 3 October 2013
pT7: RBS: His6 - SUMO: MaximinH5
This part codes for the peptide 'Maximin H5' tagged with 'His-6-SUMO' molecule. Its production is triggered by IPTG (through pT7 promoter).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 46
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 170
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 355
- 1000COMPATIBLE WITH RFC[1000]
Characterization
For more info, visit [http://2013.igem.org/Team:TU-Delft/Peptides#SUMO TU Delft iGEM13 Wiki]
Peptide production
Introduction:
In the project by iGEM 13 Team TU Delft, a SUMO-peptide fusion was opted as a suitable expression system as they make the fusion proteins more soluble. The peptide by itself is not soluble in the cytoplasm but making a fusion of peptide with Small Ubiquitin like Modifiers (SUMO) will increase the solubility of the peptide, thus increasing the cytoplasmic fraction of the peptide.
A gene was constructed in such a way that the SUMO-peptide production was driven by the strong T7 phage promoter. This gene containing plasmid was harboured in a BL21(DE3) strain that has lac promoter driven T7 polymerase. Upon induction by IPTG the SUMO peptide fusion is produced as a soluble protein fraction.
The protocol can be seen [http://2013.igem.org/Team:TU-Delft/Protocol_10#protocol_10 here].
Result:
- Colony PCR SUMO Peptide
The presence of plasmid with gene inserts encoding the SUMO-peptide (BBa_K1022101, BBa_K1022102, BBa_K1022103) was confirmed by a colony pcr. The expected size of the insert with the T7 promoter was approximately around 400 bp which was clearly evident from the agarose gel picture. An Eurogentec Smartladder MW-1700-10 (https://secure.eurogentec.com/uploads/TDS-MW-1700-10.pdf) was used to identify the size of the fragments.
- Tricine gel SUMO-Peptide
Then strains were subjected to protein expression by means of IPTG induction. The cell lysate and the centrifuged cell debris were run on a 16 % tricine gels at 90 V for 90 minutes. The figure clearly shows a over-expression of our fusion protein at ~15 kDa both in whole cell lysate (CL) and the membrane (M), This was from just 10 µg of total protein for cell lysate (CL) and 30 µg of total protein in the membrane (M).
Highlights:
The SUMO domain that was used in our project was synthesized as a gene from DNA 2.0. The SUMO was designed to be bio-brick compatible without the restriction sites that are present in the prefix and suffix. The restriction sites where removed without disturbing the codons.
The Biobrick was designed in such a way that the ease of cloning SUMO fusion protein to be expressed has been improved. An Age I restriction site was introduced at the end of SUMO which when translated to amino acids Tyrosine Glycine Glycine. This is the recognition sequence of the Ulp-1 protease that can cleave the peptides from the SUMO without any scar. So, one can use Age I and any one of the suffix enzyme to make a SUMO fusion.
MIC determination
The MIC of peptide on S. delphini, B. subtilis and E. coli was also done. This characterization can be seen in construct BBa_K1022110.
Also visit the [http://2013.igem.org/Team:TU-Delft/PeptideCharacterization Peptide characterization] page on TU Delft iGEM13 Wiki to know more about the MIC experiments.