Difference between revisions of "Part:BBa K1149020"
| Line 11: | Line 11: | ||
https://static.igem.org/mediawiki/igem.org/f/fb/PlasticityBluePlate.JPG | https://static.igem.org/mediawiki/igem.org/f/fb/PlasticityBluePlate.JPG | ||
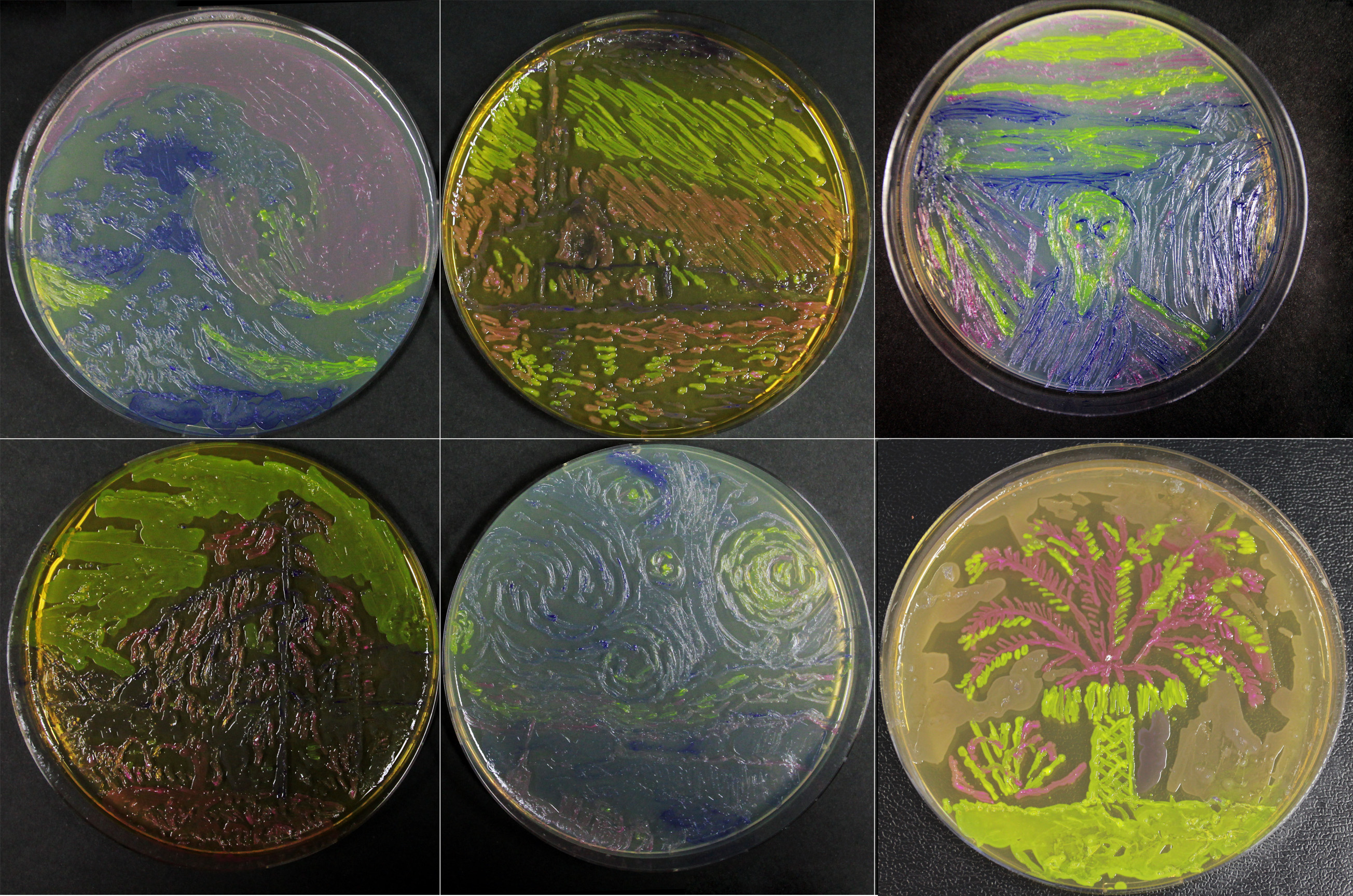
| − | In addition this, we have shown that bacterial paintings are achievable with this amazing biobrick (blue): | + | In addition to this, we have shown that bacterial paintings are achievable with this amazing biobrick (blue): |
| − | [[File:Originals_combined_small.jpg | + | [[File:Originals_combined_small.jpg|600px]] |
We have observed change in colour of an Overnight culture with the construct. However, it was not as blue as the overnight cultures obtained by Uppsala 2011. | We have observed change in colour of an Overnight culture with the construct. However, it was not as blue as the overnight cultures obtained by Uppsala 2011. | ||
Revision as of 18:41, 29 September 2013
Promoter J23104 - RBS B0034 - amilCP
The chromoprotein from the coral Acropora millepora, amilCP, naturally exhibits strong color when expressed. The protein has an absorbance maximum at 588 nm giving it a blue/purple color visible to the naked eye, thereby requiring no instruments to observe. This is a composite part from BBa_K608002 and BBa_K592009. It is functionally similar to BBa_K592031 which has a J23101 promoter instead of the J23104 but contains the same RBS and amilCP.
The blue coloration made it very easy to select the ligations with the insert from the transformation plate:

We were inspired by the beautiful blue coral colour and taught the bacteria how to write our name with it.

In addition to this, we have shown that bacterial paintings are achievable with this amazing biobrick (blue):
We have observed change in colour of an Overnight culture with the construct. However, it was not as blue as the overnight cultures obtained by Uppsala 2011.

We have verified the sequence and detected 3 mutations compared to the expected results. However, these are located after the prefix and at the scar site and do not affect the coding region or the function of the construct.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
