Difference between revisions of "Part:BBa K786031"
| Line 37: | Line 37: | ||
To improve the efficiency for construction, we split them into two biobricks containing Cas3 and the R-S-R part, and synthesize them separately Therefore, when testing the system in E. coli, transformation of the R-S-R biobrick alone is enough for characterization. | To improve the efficiency for construction, we split them into two biobricks containing Cas3 and the R-S-R part, and synthesize them separately Therefore, when testing the system in E. coli, transformation of the R-S-R biobrick alone is enough for characterization. | ||
| − | (a) ‘Repeat’ design | + | |
| + | '''(a) ‘Repeat’ design''' | ||
| + | |||
A palindrome can be observed by studying some samples of repeat sequences of CRISPR system of E. coli. The secondary structure of pre-crRNA shows that the base pairing helps to form a loop, which may contribute to the Cas3 and Cascade in expression and interference stages of type-I CRISPR system. | A palindrome can be observed by studying some samples of repeat sequences of CRISPR system of E. coli. The secondary structure of pre-crRNA shows that the base pairing helps to form a loop, which may contribute to the Cas3 and Cascade in expression and interference stages of type-I CRISPR system. | ||
| Line 45: | Line 47: | ||
We also tried other tools for repeat design. An online CRISPR database CRISPRdb (http://crispr.u-psud.fr/) as recommended by 2011 USC iGEM team provides repeat consensus of different strains [4]. | We also tried other tools for repeat design. An online CRISPR database CRISPRdb (http://crispr.u-psud.fr/) as recommended by 2011 USC iGEM team provides repeat consensus of different strains [4]. | ||
| − | (b) Spacer Design | + | |
| + | '''(b) Spacer Design''' | ||
| + | |||
PAM (Proto-spacer Adjacent Motif) is located adjacent to the proto-spacer, upstream for a sense strand or downstream for an antisense strand. This 3-bp motif is essential for the proto-spacer recognition by Cascade complex. In E. coli, Cascade tolerates at least four PAMs, CTT, CAT, CTC and CCT in an antisense strand [5]. | PAM (Proto-spacer Adjacent Motif) is located adjacent to the proto-spacer, upstream for a sense strand or downstream for an antisense strand. This 3-bp motif is essential for the proto-spacer recognition by Cascade complex. In E. coli, Cascade tolerates at least four PAMs, CTT, CAT, CTC and CCT in an antisense strand [5]. | ||
| Line 55: | Line 59: | ||
| + | |||
| + | ---- | ||
| + | |||
| + | == References: == | ||
| + | [1] Makarova KS, Haft DH, Barrangou R, et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 9: 467-477. | ||
| + | |||
| + | [2] Brouns SJ, Jore MM, Lundgren M, et al. (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 321: 960-964. | ||
| + | |||
| + | [3] Yosef I, Goren MG, Qimron U (2012). Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40: 5569-5576. | ||
| + | |||
| + | [4] Grissa I, Vergnaud G, Pourcel C (2007). The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 8: 172. | ||
| + | |||
| + | [5] Semenova E, Jore MM, Datsenko KA, et al. (2011). Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 108: 10098-10103. | ||
| + | |||
| + | [6] Westra ER, van Erp PB, Kunne T, et al. (2012). CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 46: 595-605. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 14:55, 28 September 2012
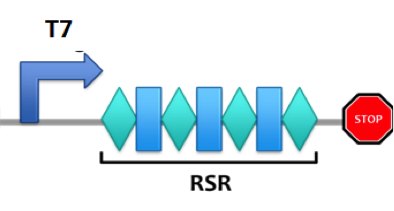
T7promoter-RSR-T7teminator
It is a repeat-spacer-repeat region designed according to E.coli's CRISPR/Cas system. It also includes a T7 promoter and T7 terminator. THe spacer is designed to target on a segment of non-coding-sequence in E.coli's genome. This biobrick can be transformed into E.coli cells to hopefully construct a suicide system with E.coli's CRISPR/Cas system.
Introduction: New Safety Approach
Multidrug resistance on microorganism has already changed from nightmare to reality. Horizontal gene transfer is one of the ways for bacteria to obtain exogenous genetic material. The previous proposal on biosafety of synthetic biology by other iGEM teams have led to achievements on different approach, such as incorporating active inhibition of horizontal gene transfer (Imperial 2011), using antibiotic-free selection to reduce potential multidrug resistance on antibiotic, and labeling spec sheet information on engineered bacteria for identity (CUHK 2010).
It is well-known that DNA molecule can be long lasting for thousands years. Therefore, even the engineered bacteria was killed after experiments, the artificial genetic material such as synthetic DNA can still present in the environment. Other bacteria can uptake these genetic information and possibly incorporate into their genome. The active uptake mechanism of DNA from the environment to bacteria can be known as “natural genetic transformation”. Thus, this intelligent design can bypass the previous proposed biosafety measures.
Herein, our proposed system allows a selective cleavage of the target gene, such as antibiotic gene and biobrick gene, by activating an exonuclease to recognize the specific region and undergo digestion. This approach allows regulation of the engineered microorganism down from DNA level.
I. Mechanism: CRISPR/Cas systems
CRISPR/Cas systems are the adaptive immunity systems that are present in many archaea and bacteria to protect themselves from invading genetic materials such as viral DNA. There are three types of CRISPR/Cas system. Here we only exploit type I-E from Escherichia coli (E. coli) to focus on expression and interference stage. This system presents potential in developing into a novel tool in order to reach higher safety standard of using engineered microorganism machine.
Foreign double strand DNA (dsDNA) will be targeted by the spacers from repeat-spacer region (RSR) and destroyed by one of the CRISPR/Cas components, Cas3. Through engineering the spacer sequence, theoretically we can target any dsDNA of sequence complementary to the spacer(s) [1].
By re-designing the targeting sequence, we can cleave the transformed plasmid in certain bacteria to reduce environmental hazard, or even manage to create a controlled suicide system to reduce leakage of genetic information from the bacteria. Thus our system provides an alternative to control safety level of engineered microorganism machine.
II.Biobrick design
Our biobrick is designed to be an integration of two parts, the protein part and the R-S-R part. Sequences of proteins Cas3 and cascade complex subunits are located in the upstream of the RSR system.
The number of the repeat and spacer are not fixed and the distance from the promoter does not affect the functionality.
The protein part in the biobrick is not essential for the system to work, as the type I-E CRISPR/Cas system is an endogenous system in E. coli. Since this part is designed to improve the sufficiency of system functionality, we choose to use Pveg, a strong constitutive promoter. This choice also makes it possible to utilize this system in other microorganisms where CRISPR system does not exist naturally, such as Bacillus subtilis.
To improve the efficiency for construction, we split them into two biobricks containing Cas3 and the R-S-R part, and synthesize them separately Therefore, when testing the system in E. coli, transformation of the R-S-R biobrick alone is enough for characterization.
(a) ‘Repeat’ design
A palindrome can be observed by studying some samples of repeat sequences of CRISPR system of E. coli. The secondary structure of pre-crRNA shows that the base pairing helps to form a loop, which may contribute to the Cas3 and Cascade in expression and interference stages of type-I CRISPR system.
From related research papers [2, 3], we obtained several repeat sequences of E. coli strain K-12. Although repeat sequences from different sources may not be exactly identical, similar characteristics can be found.
We also tried other tools for repeat design. An online CRISPR database CRISPRdb (http://crispr.u-psud.fr/) as recommended by 2011 USC iGEM team provides repeat consensus of different strains [4].
(b) Spacer Design
PAM (Proto-spacer Adjacent Motif) is located adjacent to the proto-spacer, upstream for a sense strand or downstream for an antisense strand. This 3-bp motif is essential for the proto-spacer recognition by Cascade complex. In E. coli, Cascade tolerates at least four PAMs, CTT, CAT, CTC and CCT in an antisense strand [5].
Seed sequence, as indicated in the graph, is another essential for spacer design. Mutants on seed sequence may lead to “escape”. Thus for these 7 base pairs, the crRNA must complement to the target DNA. In our design, we have the whole spacer to be completely complementary to the protospacer, so does the crRNA. Therefore, the problem of "escape" due to the mismatch in seed sequence is not crucial.
The designed length of spacer in our CRISPR system should be 32bp. The spacer is specially desigened to target on the genomic DNA of a typical strain of E. coli, K12 MG1655. It is a non-coding DNA sequence since it is important to confirm that the factor causing the death of E. coli is due to the cleavage of its genomic DNA but not the malfunction of the gene being targeted, so a non-coding DNA sequence is preferred than a coding DNA sequence. The selection of the non-coding DNA sequence of E. coli K12 MG1655 is random, and the 32 bp is taken from 121595-121626 (GenBank: U00096.2 )

References:
[1] Makarova KS, Haft DH, Barrangou R, et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 9: 467-477.
[2] Brouns SJ, Jore MM, Lundgren M, et al. (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 321: 960-964.
[3] Yosef I, Goren MG, Qimron U (2012). Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40: 5569-5576.
[4] Grissa I, Vergnaud G, Pourcel C (2007). The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 8: 172.
[5] Semenova E, Jore MM, Datsenko KA, et al. (2011). Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 108: 10098-10103.
[6] Westra ER, van Erp PB, Kunne T, et al. (2012). CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 46: 595-605.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 816
- 1000COMPATIBLE WITH RFC[1000]