Difference between revisions of "Part:BBa K3457009"
| Line 1: | Line 1: | ||
| − | + | ==iGEM 2020 QHFZ-China, new documentation (For Bronze)== | |
| − | + | <h3><b>Group: QHFZ-China iGEM 2020</b></h3> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | < | + | |
<h3><b>Author: Yixian Yang</b></h3> | <h3><b>Author: Yixian Yang</b></h3> | ||
| − | < | + | <p> We measured [https://parts.igem.org/Part:BBa_J23100 BBa_J23100], [https://parts.igem.org/Part:BBa_J23107 BBa_J23107] and [https://parts.igem.org/Part:BBa_J23109 BBa_J23109] as a strong, moderate and weak promoter respectively in 2020. For all the experiments below, we use <i>E. coli</i> BL21(DE3) strain.</p> |
| − | [[File:T--QHFZ-China-- | + | <h3>Part 1: Measurement with a reprter, sfGFP</h3> |
| − | + | <h4>Description</h4> | |
| − | <p style="clear:left;"> | + | <p> First, we measured the strength of the promoter by sfGFP [https://parts.igem.org/Part:BBa_K3457015 BBa_K3457015].</p> |
| − | + | <h4>Protocol</h4> | |
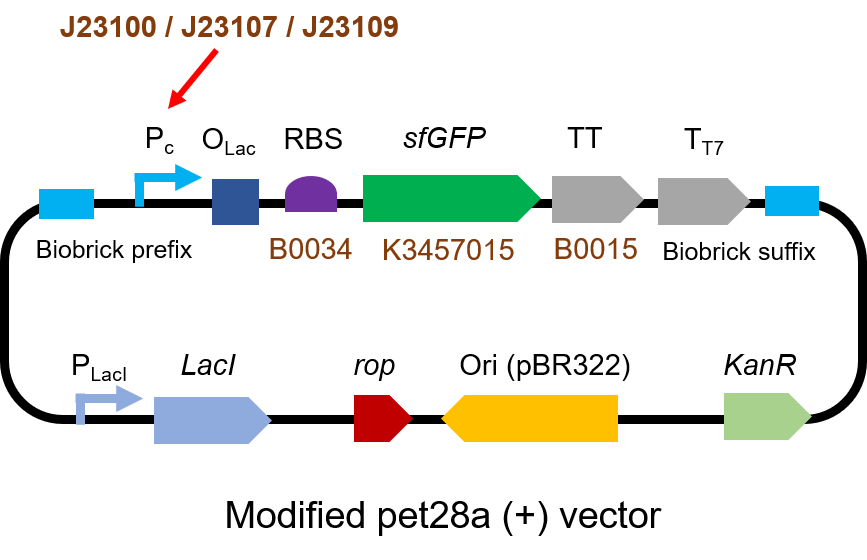
| − | < | + | <p> The gene circuit we used is as below:</p> |
| − | <h4>< | + | [[File:T--QHFZ-China--J2310-1.png|600px|thumb|left|Figure 1. The Schematic cartoon of the DNA construct to test J23100 / |
| − | <p style="clear:left;"> | + | J23107 / J23109 with sfGFP.]] |
| − | + | <p style="clear:left;"> The protocol is as below: <br> | |
| − | < | + | (1) Pick clones which are in good condition and put them into 500 μL LB medium containing antibiotics. Shake them to |
| − | <p | + | grow at 37℃ for 5~7 hours until the bacteria solution becomes turbid. <br> |
| − | + | (2) Add 2mM iPTG into 3 mL LB medium containing antibiotics. Add 3 μL of the bacteria solution mentioned in step 1 | |
| − | [[File:T--QHFZ-China-- | + | to dilute the bacteria by the ratio of 1:1000. Shake the solution to grow the bacteria at 37℃ overnight.<br> |
| − | + | (3) The bacteria solution was centrifuged and the LB medium was removed. Then the bacteria was resuspended by PBS. | |
| − | <p style="clear:left;"> | + | 100 μL such solution was put into a well of a 96-well palte. The GFP fluorescence and OD<sub>600</sub> were detected |
| − | + | by a microplate readers (Bio-Teck). The parameters are: exciting light: 488 nm, light reception: 520 nm, gain: 50. | |
| − | [[File:T--QHFZ-China--freeze-dry protocol.jpg| | + | <br> |
| − | + | (4) The value of PBS was deducted from the result above. GFP / OD<sub>600</sub> was calculated.<br> | |
| + | </p> | ||
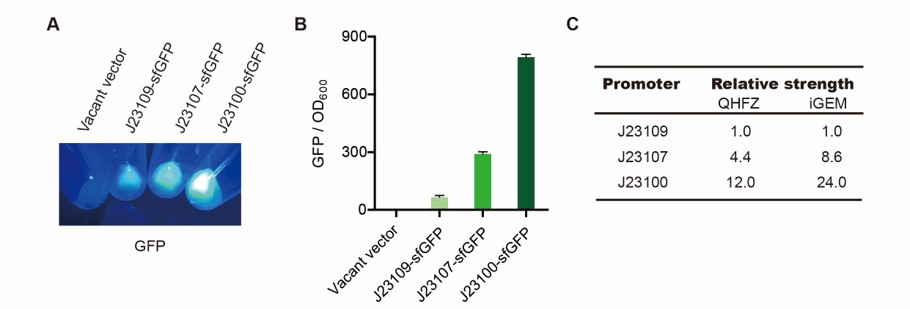
| + | <h4>Result</h4> | ||
| + | [[File:T--QHFZ-China--sfGFP.jpg|600px|thumb|left|Figure 2. sfGFP was expressed with J23100 / J23107 / J23109.]] | ||
| + | <p style="clear:left;"> We set the strehgth of J23109 as 1. The relative strengths of J23107 and J23109 were 4.4 and | ||
| + | 12.0. Though they are not the same as the data at the top of this page, they worked well anb the strength order of | ||
| + | the three promoters was accordance was consistent with other people's data. The difference may owe to the certain | ||
| + | gene circuit and protocol. </p> | ||
| + | <h3>Part 2: Measurement with CHAS 106094</h3> | ||
| + | <h4>Description</h4> | ||
| + | <p> Second, we measured the strength of the promoter by CAHS 106094 | ||
| + | [https://parts.igem.org/Part:BBa_K3457012 BBa_K3457012]. This year, we used CAHS 106094 to protect bacteria from | ||
| + | freeze-drying and dry storage. We used different promoters to adjust the expression level of CAHS 106094, to study | ||
| + | the relationship between the survival rate and CAHS 106094 expression level.</p> | ||
| + | <h4>Protocol</h4> | ||
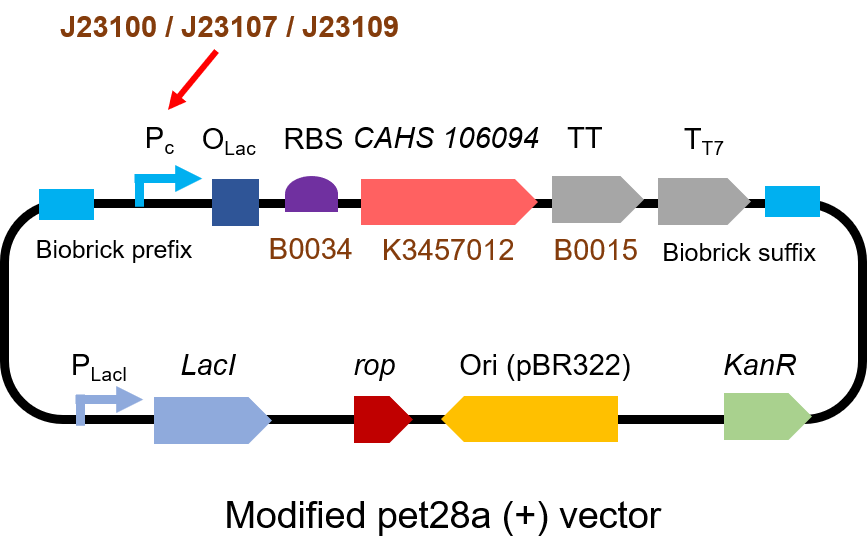
| + | <p> The gene circuit we used is as below:</p> | ||
| + | [[File:T--QHFZ-China--J2310-2.png|600px|thumb|left|Figure 3. The Schematic cartoon of the DNA construct to test J23100 / | ||
| + | J23107 / J23109 with sfGFP.]] | ||
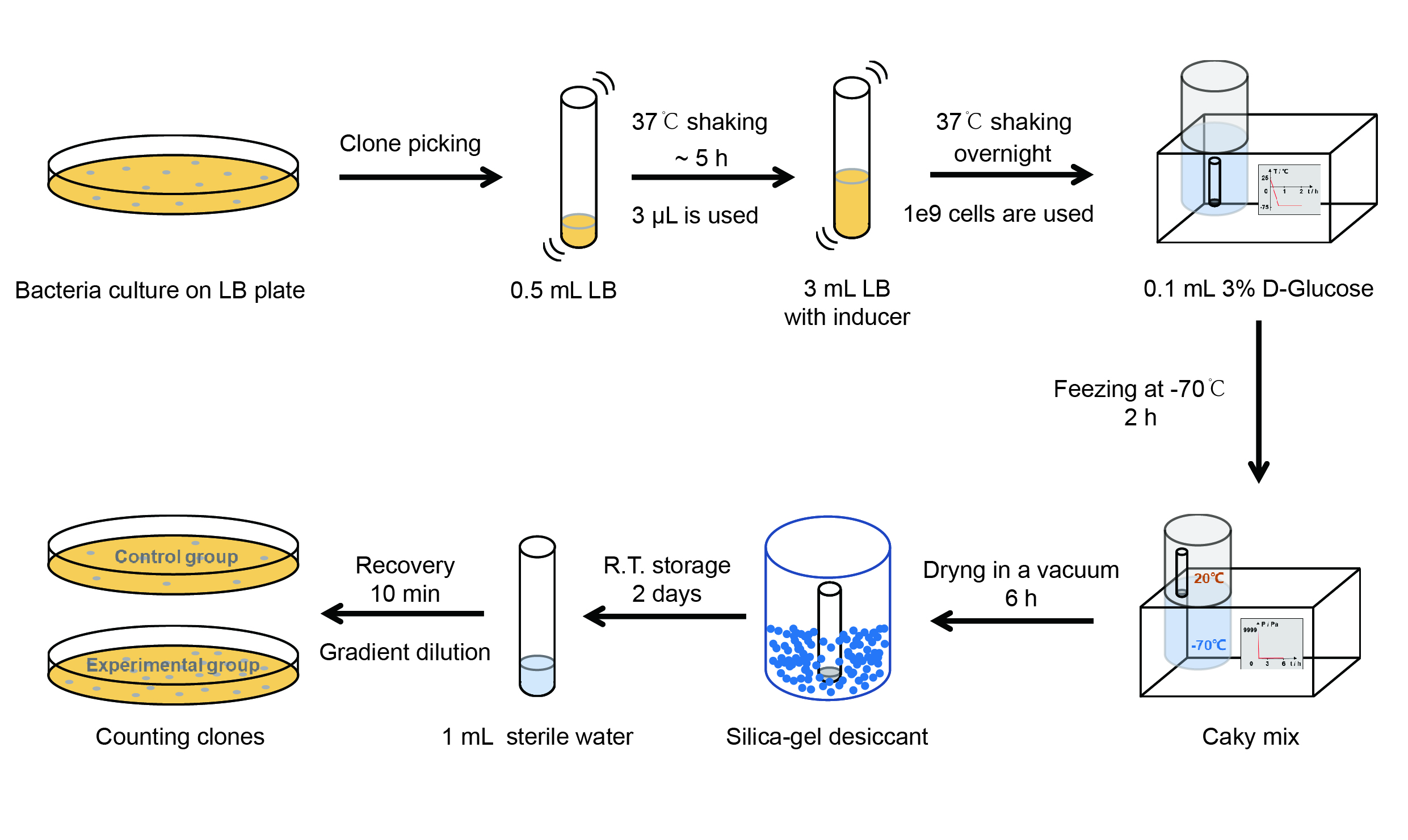
| + | <p style="clear:left;"> The protocol is as below: </p> | ||
| + | [[File:T--QHFZ-China--freeze-dry protocol.jpg|600px|thumb|left|Figure 4. Experiment protocol.]] | ||
<p style="clear:left;"> | <p style="clear:left;"> | ||
| − | 【Day 1】Induction culture<br> | + | 【Day 1】Induction culture<br> |
| − | (1) Pick clones which are in good condition and put them into 500 μL LB medium containing antibiotics. Shake them to grow at 37℃ for 5~7 hours until the bacteria solution becomes turbid. <br> | + | (1) Pick clones which are in good condition and put them into 500 μL LB medium containing antibiotics. Shake them to |
| − | (2) Add 2mM iPTG into 3 mL LB medium containing antibiotics. Add 3 μL of the bacteria solution mentioned in step 1 to dilute the bacteria by the ratio of 1:1000. Shake the solution to grow the bacteria at 37℃ overnight.<br> | + | grow at 37℃ for 5~7 hours until the bacteria solution becomes turbid. <br> |
| − | 【Day 2】Freeze-dried<br> | + | (2) Add 2mM iPTG into 3 mL LB medium containing antibiotics. Add 3 μL of the bacteria solution mentioned in step 1 |
| − | (1) If fluorescence induced by the iPTG is detectable in the control group (GFP), continue conducting the experiment.<br> | + | to dilute the bacteria by the ratio of 1:1000. Shake the solution to grow the bacteria at 37℃ overnight.<br> |
| − | (2) Use spectrophotometer to measure the OD<sub>600</sub> of the bacteria solution, OD<sub>600</sub> = 1 equals to 10<sup>9</sup> cells. If the OD<sub>600</sub> value is between 0.1 and 1, There is a linear relationship between OD<sub>600</sub> and bacterial density. Calculate the volume of bacterial solution for 10<sup>9</sup> cells by using the formula V = 100 / (OD<sub>600</sub> × Dilution ratio).<br> | + | 【Day 2】Freeze-dried<br> |
| − | (3) Take out a measured amount of 10<sup>9</sup> cells and centrifuge it at 8000 rpm for 3 min. Then pour out the supernatant.<br> | + | (1) If fluorescence induced by the iPTG is detectable in the control group (GFP), continue conducting the |
| − | (4) Resuspend the bacteria in a 15 mL tube with pre-refrigerated 100 μL 3% glucose solution.<br> | + | experiment.<br> |
| − | (5) Take off the cover of the tube and put the bacteria into the cold trap. Open the compressor of the lyophilization machine and freeze the shake tube for 2 h at -70℃.<br> | + | (2) Use spectrophotometer to measure the OD<sub>600</sub> of the bacteria solution, OD<sub>600</sub> = 1 equals to |
| − | (6) Put the caky bacteria solution into the drying chamber of the lyophilization machine. Open the vacuum pump to dry it in vacuum for 6h at 1 Pa vacuum degree.<br> | + | 10<sup>9</sup> cells. If the OD<sub>600</sub> value is between 0.1 and 1, There is a linear relationship between |
| − | (7) Turn off the vacuum pump, place it at seal box filled with silica-gel desiccant a for 2 days at room temperature.<br> | + | OD<sub>600</sub> and bacterial density. Calculate the volume of bacterial solution for 10<sup>9</sup> cells by using |
| − | 【Day 3】Room temperature storage<br> | + | the formula V = 100 / (OD<sub>600</sub> × Dilution ratio).<br> |
| − | 【Day 4】Detect the survival rate<br> | + | (3) Take out a measured amount of 10<sup>9</sup> cells and centrifuge it at 8000 rpm for 3 min. Then pour out the |
| − | (1) Add 1 mL of sterile water to the tube, vortex for 15 s, placed it at room temperature for 10 min.<br> | + | supernatant.<br> |
| − | (2) Adjust the density of the bacteria solution by gradient dilution, then spread 100 μL of the bacteria solution on the LB plate.<br> | + | (4) Resuspend the bacteria in a 15 mL tube with pre-refrigerated 100 μL 3% glucose solution.<br> |
| − | (3) If the density above is not suitable, take 100μL of the solution and spread it on the LB plate after several gradient dilutions.<br> | + | (5) Take off the cover of the tube and put the bacteria into the cold trap. Open the compressor of the |
| − | (4) Culture the bacteria overnight at 37℃.<br> | + | lyophilization machine and freeze the shake tube for 2 h at -70℃.<br> |
| − | 【Day 5】Cell Count<br> | + | (6) Put the caky bacteria solution into the drying chamber of the lyophilization machine. Open the vacuum pump to |
| − | (1) Take out the LB plate and take photos to record experimental results.<br> | + | dry it in vacuum for 6h at 1 Pa vacuum degree.<br> |
| − | (2) Use the automatic cell counting function of Image J to count the colone number on the LB plate, then compare the results between each group. | + | (7) Turn off the vacuum pump, place it at seal box filled with silica-gel desiccant a for 2 days at room |
| + | temperature.<br> | ||
| + | 【Day 3】Room temperature storage<br> | ||
| + | 【Day 4】Detect the survival rate<br> | ||
| + | (1) Add 1 mL of sterile water to the tube, vortex for 15 s, placed it at room temperature for 10 min.<br> | ||
| + | (2) Adjust the density of the bacteria solution by gradient dilution, then spread 100 μL of the bacteria solution on | ||
| + | the LB plate.<br> | ||
| + | (3) If the density above is not suitable, take 100μL of the solution and spread it on the LB plate after several | ||
| + | gradient dilutions.<br> | ||
| + | (4) Culture the bacteria overnight at 37℃.<br> | ||
| + | 【Day 5】Cell Count<br> | ||
| + | (1) Take out the LB plate and take photos to record experimental results.<br> | ||
| + | (2) Use the automatic cell counting function of Image J to count the colone number on the LB plate, then compare the | ||
| + | results between each group.<br> | ||
</p> | </p> | ||
| + | <h4>Result</h4> | ||
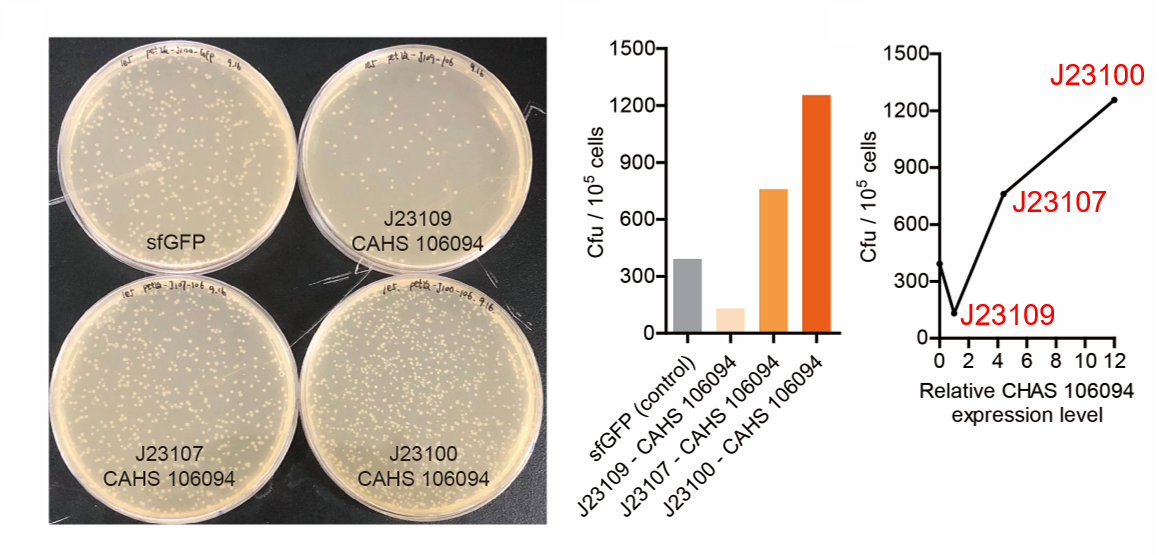
| + | [[File:T--QHFZ-China--J2310-3.png|600px|thumb|left|Figure 5. The Cfu of bacteria expressing CAHS 106094 after | ||
| + | freeze-drying with J23100 / J23107 / J23109.]] | ||
| + | <p style="clear:left;"> As expected, J23100 is the strongest promoter and it gave the best survival rate. J23107 is | ||
| + | the second and J23109 seemed too weak to express enough CAHS 106094. In conclusion, J23100 and J23107 is effective | ||
| + | in this situation, but J23109 is not.</p> | ||
| + | <!-- The end of QHFZ-China 2020--> | ||
| − | < | + | <span id="UT_Austin_2019"><br><br><br><br><br></span> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | < | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Revision as of 08:49, 26 October 2020
Contents
iGEM 2020 QHFZ-China, new documentation (For Bronze)
Group: QHFZ-China iGEM 2020
Author: Yixian Yang
We measured BBa_J23100, BBa_J23107 and BBa_J23109 as a strong, moderate and weak promoter respectively in 2020. For all the experiments below, we use E. coli BL21(DE3) strain.
Part 1: Measurement with a reprter, sfGFP
Description
First, we measured the strength of the promoter by sfGFP BBa_K3457015.
Protocol
The gene circuit we used is as below:
The protocol is as below:
(1) Pick clones which are in good condition and put them into 500 μL LB medium containing antibiotics. Shake them to
grow at 37℃ for 5~7 hours until the bacteria solution becomes turbid.
(2) Add 2mM iPTG into 3 mL LB medium containing antibiotics. Add 3 μL of the bacteria solution mentioned in step 1
to dilute the bacteria by the ratio of 1:1000. Shake the solution to grow the bacteria at 37℃ overnight.
(3) The bacteria solution was centrifuged and the LB medium was removed. Then the bacteria was resuspended by PBS.
100 μL such solution was put into a well of a 96-well palte. The GFP fluorescence and OD600 were detected
by a microplate readers (Bio-Teck). The parameters are: exciting light: 488 nm, light reception: 520 nm, gain: 50.
(4) The value of PBS was deducted from the result above. GFP / OD600 was calculated.
Result
We set the strehgth of J23109 as 1. The relative strengths of J23107 and J23109 were 4.4 and 12.0. Though they are not the same as the data at the top of this page, they worked well anb the strength order of the three promoters was accordance was consistent with other people's data. The difference may owe to the certain gene circuit and protocol.
Part 2: Measurement with CHAS 106094
Description
Second, we measured the strength of the promoter by CAHS 106094 BBa_K3457012. This year, we used CAHS 106094 to protect bacteria from freeze-drying and dry storage. We used different promoters to adjust the expression level of CAHS 106094, to study the relationship between the survival rate and CAHS 106094 expression level.
Protocol
The gene circuit we used is as below:
The protocol is as below:
【Day 1】Induction culture
(1) Pick clones which are in good condition and put them into 500 μL LB medium containing antibiotics. Shake them to
grow at 37℃ for 5~7 hours until the bacteria solution becomes turbid.
(2) Add 2mM iPTG into 3 mL LB medium containing antibiotics. Add 3 μL of the bacteria solution mentioned in step 1
to dilute the bacteria by the ratio of 1:1000. Shake the solution to grow the bacteria at 37℃ overnight.
【Day 2】Freeze-dried
(1) If fluorescence induced by the iPTG is detectable in the control group (GFP), continue conducting the
experiment.
(2) Use spectrophotometer to measure the OD600 of the bacteria solution, OD600 = 1 equals to
109 cells. If the OD600 value is between 0.1 and 1, There is a linear relationship between
OD600 and bacterial density. Calculate the volume of bacterial solution for 109 cells by using
the formula V = 100 / (OD600 × Dilution ratio).
(3) Take out a measured amount of 109 cells and centrifuge it at 8000 rpm for 3 min. Then pour out the
supernatant.
(4) Resuspend the bacteria in a 15 mL tube with pre-refrigerated 100 μL 3% glucose solution.
(5) Take off the cover of the tube and put the bacteria into the cold trap. Open the compressor of the
lyophilization machine and freeze the shake tube for 2 h at -70℃.
(6) Put the caky bacteria solution into the drying chamber of the lyophilization machine. Open the vacuum pump to
dry it in vacuum for 6h at 1 Pa vacuum degree.
(7) Turn off the vacuum pump, place it at seal box filled with silica-gel desiccant a for 2 days at room
temperature.
【Day 3】Room temperature storage
【Day 4】Detect the survival rate
(1) Add 1 mL of sterile water to the tube, vortex for 15 s, placed it at room temperature for 10 min.
(2) Adjust the density of the bacteria solution by gradient dilution, then spread 100 μL of the bacteria solution on
the LB plate.
(3) If the density above is not suitable, take 100μL of the solution and spread it on the LB plate after several
gradient dilutions.
(4) Culture the bacteria overnight at 37℃.
【Day 5】Cell Count
(1) Take out the LB plate and take photos to record experimental results.
(2) Use the automatic cell counting function of Image J to count the colone number on the LB plate, then compare the
results between each group.
Result
As expected, J23100 is the strongest promoter and it gave the best survival rate. J23107 is the second and J23109 seemed too weak to express enough CAHS 106094. In conclusion, J23100 and J23107 is effective in this situation, but J23109 is not.