Difference between revisions of "Part:BBa K3140010"
| Line 19: | Line 19: | ||
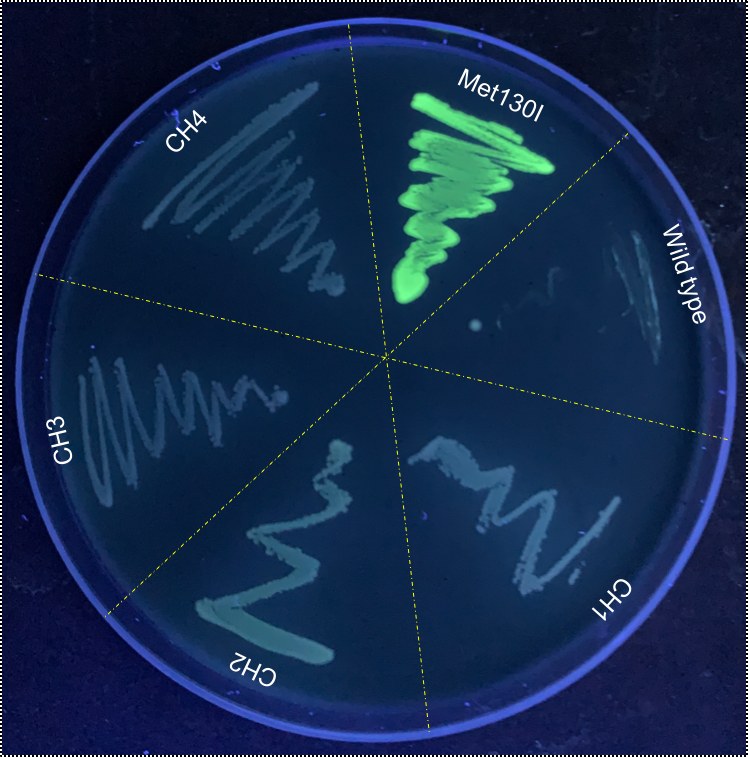
[[Image:T--Sydney_Australia--flouro_plate.png|frame|none|'''Fig. 3''': VVD36-C73A-CH4-M130I compared to VVD36-C73A, VVD36-C73A-CH1, VVD36-C73A-CH2, VVD36-C73A-CH3, and VVD36-C73A-CH4 under long-wave UV.]] | [[Image:T--Sydney_Australia--flouro_plate.png|frame|none|'''Fig. 3''': VVD36-C73A-CH4-M130I compared to VVD36-C73A, VVD36-C73A-CH1, VVD36-C73A-CH2, VVD36-C73A-CH3, and VVD36-C73A-CH4 under long-wave UV.]] | ||
| + | |||
| + | In order to gauge the thermal stability of VVD36-C73A-CH4-M130I, we conducted a melting curve analysis with comparison to sfGFP ([https://parts.igem.org/Part:BBa_I746916 BBa_I746916]). We prepared 50 mL overnight cultures of pK18:sfGFP and pK18:VVD36-C73A-CH4-M130I ([https://parts.igem.org/Part:BBa_K3140010 BBa_K3140010]). Following culture, cells were pelleted by centrifugation and resuspended in 1mL of Zietkiewicz buffer (40mM Tris-HCl, pH 7.8; 50mM NaCl: 20mM KCl; 20mM MgCl2; 5mM b-mercapto-ethanol; 10% glycerol). Cells were then lysed through bead-beating, and then centrifuged to remove supernatant. Serial dilutions of cell lysate were prepared in a 96-well PCR plate. We then used a qPCR machine with a 6-carboxy-fluorescein (FAM) filter to measure excitation and emission at 494 and 518 nm, respectively, at an initial temperature of 60°C for one minute before increasing by 0.3 degrees per second up to 95°C. | ||
| + | |||
| + | Results are summarised in '''Fig. 4'''. | ||
| + | |||
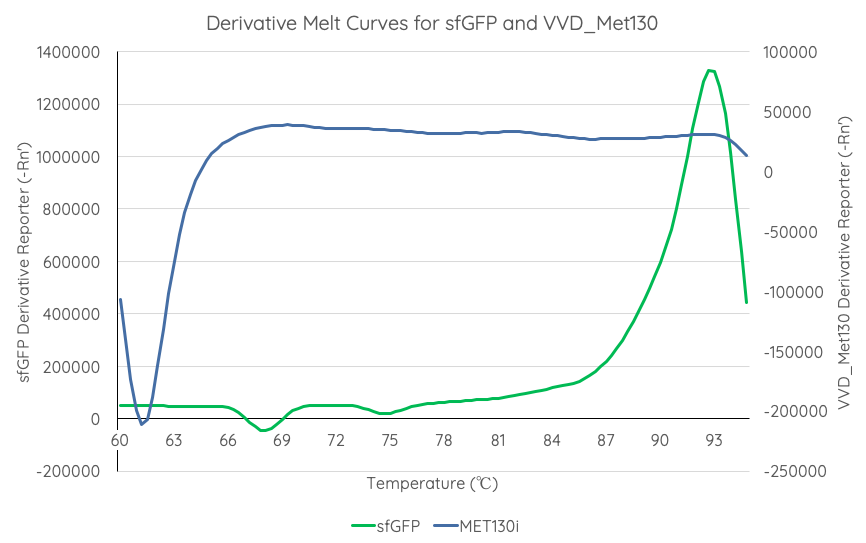
| + | [[Image:T--Sydney_Australia--sfGFPVVD_Met130DerivativeMeltCurve.png|frame|none|'''Fig 4''': We measured the thermal stability of sfGFP using the appliedbiosystems StepOnePlus Real Time PCR System using a FAM filter (max nm absorbtion 494, max emission 518) over a temperature range of 60-95°C increasing at 0.3°C per second. Temperature (°C) is displayed on the x axis, and d(Fluor)/dT is displayed on the y axis.]] | ||
| + | |||
<span class='h3bb'>'''Sequence and Features'''</span> | <span class='h3bb'>'''Sequence and Features'''</span> | ||
Revision as of 13:16, 21 October 2019
VVD36-C73A-CH4-M130I
VVD36-C73A-CH4-M103I is a fluoroprotein derived from the VIVID (VVD) blue-light photoreceptor in Neurospora crassa. A single methionine to isoleucine residue substitution in VVD36-C73A-CH4 led to this part.
Usage and Biology
VIVID (VVD) is a blue-light sensing photoreceptor from the ascomycete (spore-shooting fungus) N. crassa. It is a member of a family of proteins containing a light-oxygen-voltage-sensing (LOV) domain, which modulate circadian responses to environmental stimuli[1]. Mutation of the highly-conserved LOV domain cystine residue (Cys73) to alanine will convert VVD into a fluoroprotein. In addition, previous work [2] indicates that truncation of the first 36 amino acids of VVD increases its stability in heterologous systems. Our VVD part incorporates both of these changes.
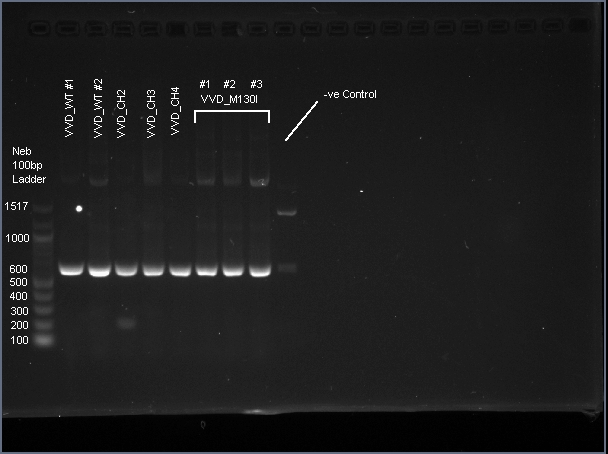
VVD36-C73A-CH4-M103I is a 456 bp sequence, encoding a 151 amino acid peptide (17.2 kDa). We cloned VVD36-C73A, VVD36-C73A-CH1, VVD36-C73A-CH2, VVD36-C73A-CH3, and VVD36-C73A-CH4 into pK18, which were then transformed into Escherichia coli TOP10 cells. In order to validate the insertion of these genes, we conducted a PCR with primers that were specific to the pK18 backbone, which should a 624 bp product. This was verified with agarose gel electrophoresis, demonstrating had the expected size (Fig. 1).
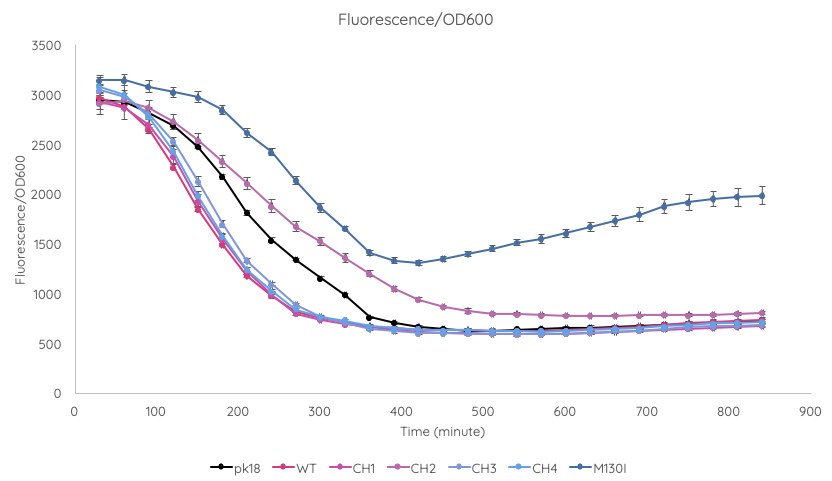
We conducted a fluorescence assay using a TECAN Spark microplate reader Escherichia coli TOP10 cultures containing VVD36-C73A, VVD36-C73A-CH1, VVD36-C73A-CH2, VVD36-C73A-CH3, VVD36-C73A-CH4, and VVD36-C73A-CH4-M130I in pK18 (Fig. 2). As shown, VVD36-C73A-CH4-M130I exhibits the greatest fluorescence, followed by VVD36-C73A-CH2. All other VVD38-C73A variants were only slightly more fluorescent than control.
When these cultures were plated, it was immediately apparently that VVD36-C73A-CH4-M130I had strong fluorescence compared to the other VVD36-C73A variants (Fig. 3).
In order to gauge the thermal stability of VVD36-C73A-CH4-M130I, we conducted a melting curve analysis with comparison to sfGFP (BBa_I746916). We prepared 50 mL overnight cultures of pK18:sfGFP and pK18:VVD36-C73A-CH4-M130I (BBa_K3140010). Following culture, cells were pelleted by centrifugation and resuspended in 1mL of Zietkiewicz buffer (40mM Tris-HCl, pH 7.8; 50mM NaCl: 20mM KCl; 20mM MgCl2; 5mM b-mercapto-ethanol; 10% glycerol). Cells were then lysed through bead-beating, and then centrifuged to remove supernatant. Serial dilutions of cell lysate were prepared in a 96-well PCR plate. We then used a qPCR machine with a 6-carboxy-fluorescein (FAM) filter to measure excitation and emission at 494 and 518 nm, respectively, at an initial temperature of 60°C for one minute before increasing by 0.3 degrees per second up to 95°C.
Results are summarised in Fig. 4.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]