Difference between revisions of "Part:BBa K1033933"
(→Contribution) |
|||
| Line 23: | Line 23: | ||
<li> </html>the [http://https://parts.igem.org/Part:BBa_I719005 BBa_I719005] T7 promotor<html></li> | <li> </html>the [http://https://parts.igem.org/Part:BBa_I719005 BBa_I719005] T7 promotor<html></li> | ||
</ul> | </ul> | ||
| + | <br><br> | ||
| + | |||
| + | The agar plate contains E.coli (BL21(DE3)) with a p.Cons-asPink plasmid (<partinfo>BBa_K3182100</partinfo>). The promotor is constitutive which will express AsPink and makes the colonies pink. This can be used to determine whether a ligation was successful. | ||
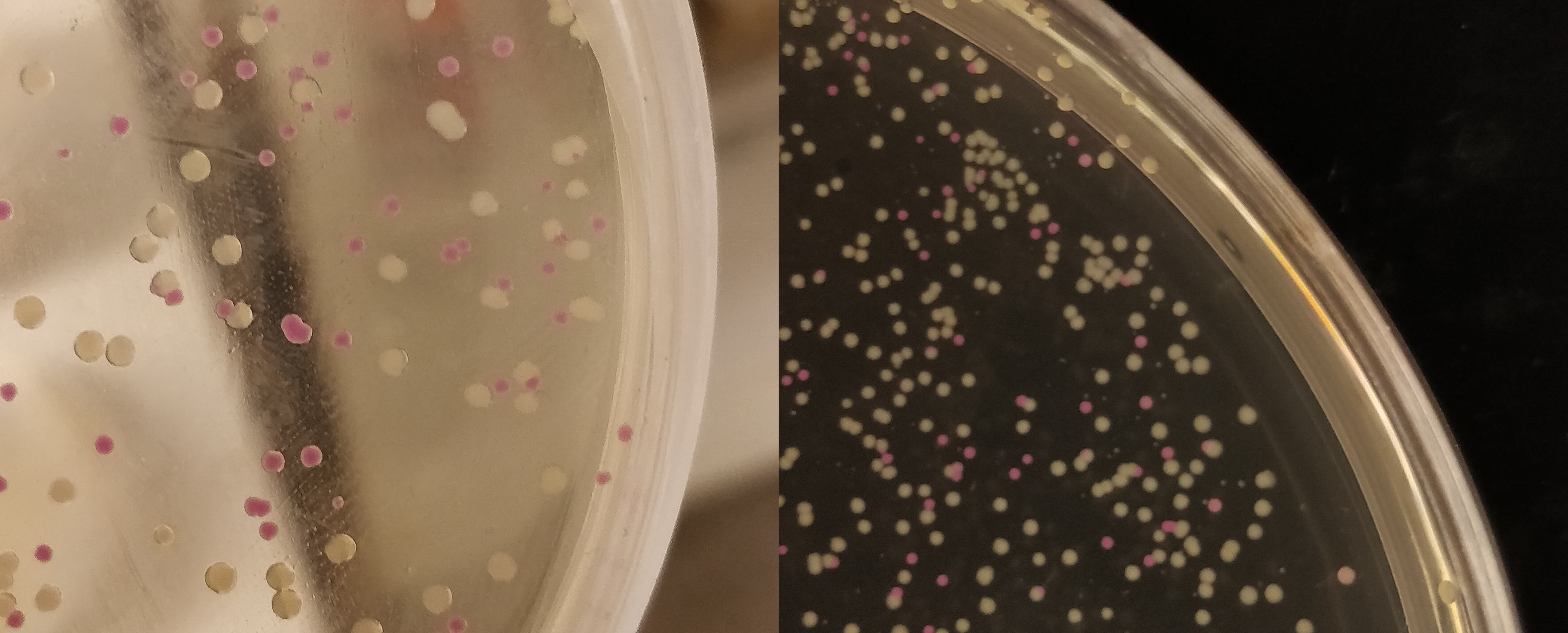
| + | </html>[[File:T--Linkoping_Sweden--pinkwhite156.jpeg|700px|thumb|left|<b>Figure 2.</b> E. coli (BL21) cells used for pink-white screening, the cells were incubated for 16 hours in 37 degrees Celsius. <partinfo>BBa_K3182100</partinfo> was cut with BamHI and PstI to remove pCons-asPink and <partinfo>BBa_K3182006</partinfo> (magainin 2) and <partinfo>BBa_K3182104</partinfo> (CHAP) was ligated into the plasmid. The white colonies indicate a successful ligation. All the colonies that were later colony screened with PCR amplification of the insert and the ampified strand was run on an agarose gel. The gel implied that all screened colonies was successful, i.e. contained <partinfo>BBa_K3182100</partinfo> with magainin 2 / CHAP instead of pCons-AsPink. ]]<html> | ||
| + | <br><br> | ||
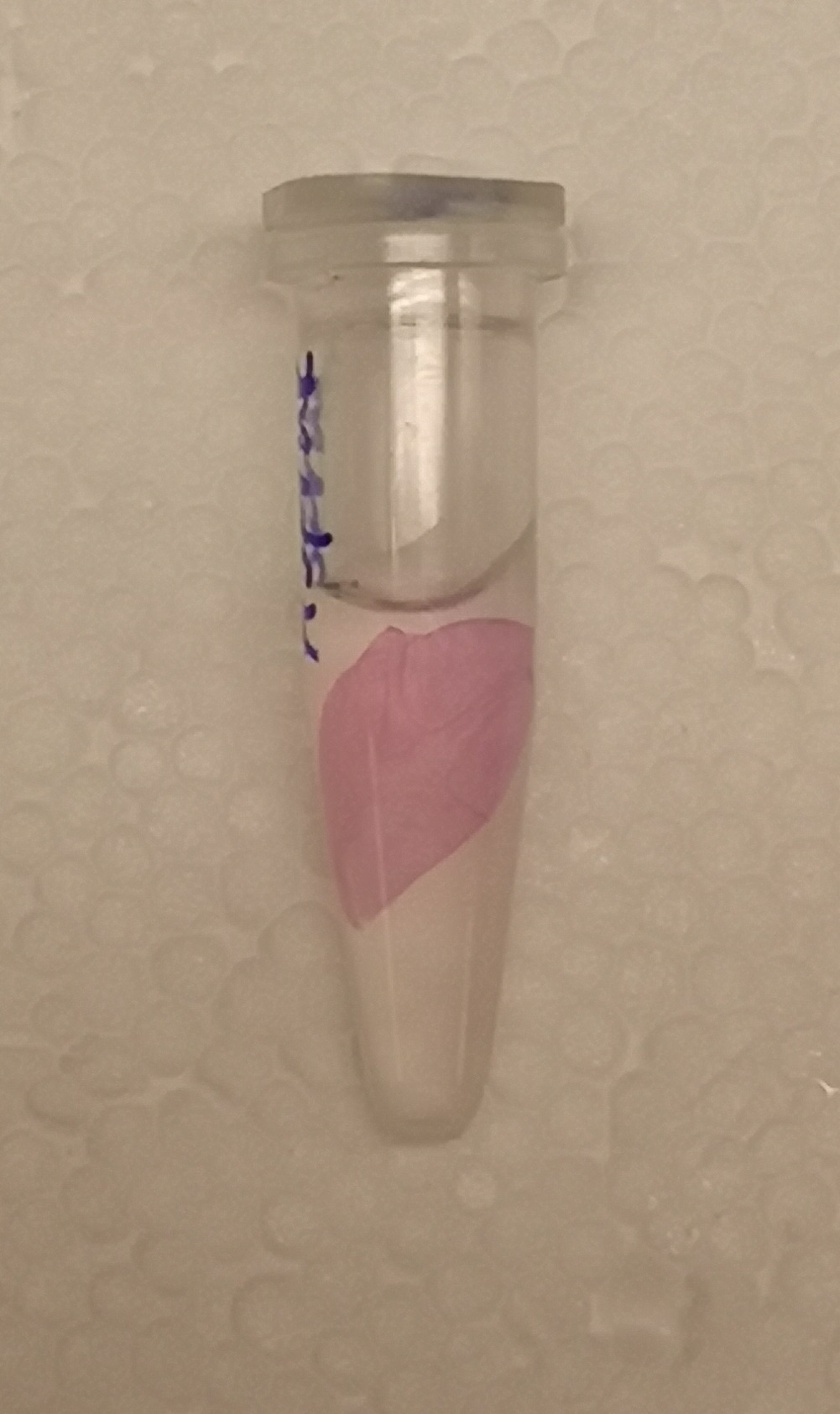
| + | </html>[[File:T--Linkoping Sweden--aspink bunden.jpeg|150px|left|thumb|<b><I>Figure 1.</I></b> Lysate (via sonication) from BL21 E. coli was incubated with Epiprotect (microbial cellulose bandage) for 1h and washed thrice with 70% ethanol.] ]] <html> | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | <br><br><br> | ||
| + | <b>CBD-asPink bindning capacity</b><br> | ||
| + | To test the bindning capacity of CBD-asPink (<partinfo>BBa_K3182000</partinfo>) the microbial cellulose bandage was suspended into sonicated lysate of BL21(DE3) with the expressed fusion protein (see <b><I>Figure 1</I></b>) and incubated for 30 minutes. | ||
| + | The bandage was then washed thrice in 70% ethanol which confirmed that the bindning of CDB-asPink was specific for the cellulose bandage.] | ||
| + | |||
| + | <br><br> | ||
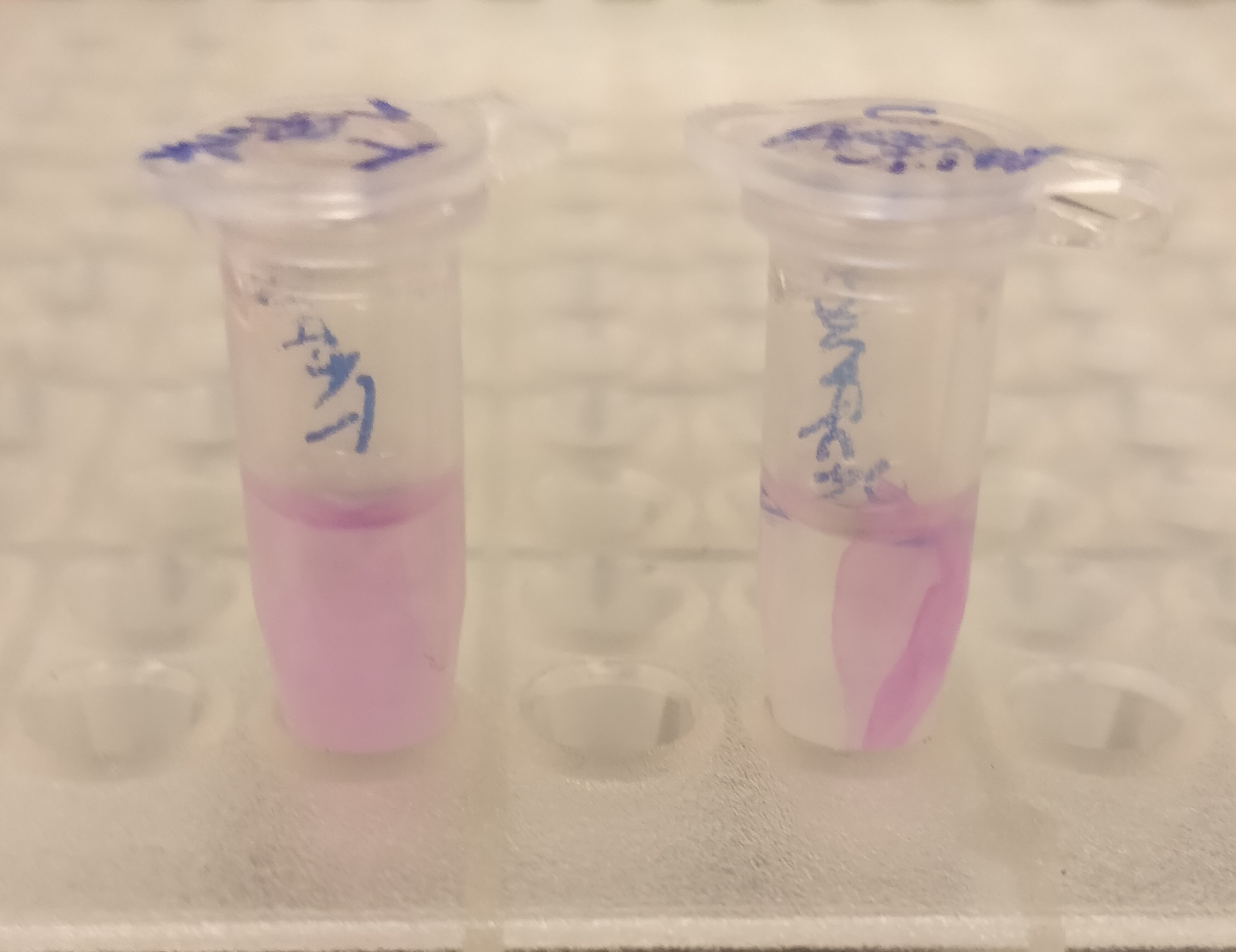
| + | </html>[[File:T--Linkoping Sweden--aspink bunden vs obunden.jpeg|150px|left|thumb|<b><I>Figure 2.</I></b> To test the release mechanism of CBD-asPink, human thrombin and cleavage buffer was added to the bandage and incubated for 16 hours on an end to end rotator. The figure is the result after the incubation with a negative control with only cleavage buffer (left) and human thrombin and cleavage buffer (right) ]] | ||
| + | <html> | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | <b> CBD-asPink with thrombin cleavage</b><br> | ||
| + | After the washes, human thrombin and cleavage buffer was added to the bandage with bound CBD-asPink to test the release mechanism of the fusion protein. The bandage was then incubated with the solution for 16 hours on an end to end rotator together with an negative control containing cellulose bandage and only cleavage buffer. | ||
| + | After the incubation, the supernatant containing thrombin was pink while the negative control containing only cleavage buffer was transparent with a clear pink cellulose bandage (see <b><I>Figure 2</I></b>). This indicates that the release mechanism work and the AsPink protein has successfully been released from the bandage into the supernatant. | ||
| + | <br><br> | ||
<br><br> | <br><br> | ||
</div> | </div> | ||
</html> | </html> | ||
| + | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
This part is useful as a reporter. | This part is useful as a reporter. | ||
Revision as of 09:43, 12 October 2019
asPink, pink chromoprotein
This chromoprotein from the coral Anemonia sulcata, asPink (also known as asCP or asFP595), naturally exhibits strong color when expressed. The protein has an absorption maximum at 572 nm giving it a pink/purple color visible to the naked eye. The strong color is readily observed in both LB or on agar plates after less than 24 hours of incubation. The protein asPink has significant sequence homologies with proteins in the GFP family.
Important: This part is not available in the registry yet, however, the same part is available from the registry with the standard RBS B0034: BBa_K1033927.
Contribution
Group: Linkoping_Sweden iGEM 2019
Author: Andreas Holmqvist and Leo Juhlin
Summary: We tested the oxygen dependency of the chromophore protein expression in E.coli BL21(DE3) cells and then measured the absorbance of the cells with an UV-spectrophotometer.
Documentation:
- the BBa_B0034 ribosome binding site
- the BBa_J23110 Constitutive promotor
- the [http://https://parts.igem.org/Part:BBa_I719005 BBa_I719005] T7 promotor
The agar plate contains E.coli (BL21(DE3)) with a p.Cons-asPink plasmid (
CBD-asPink bindning capacity
To test the bindning capacity of CBD-asPink (
CBD-asPink with thrombin cleavage
After the washes, human thrombin and cleavage buffer was added to the bandage with bound CBD-asPink to test the release mechanism of the fusion protein. The bandage was then incubated with the solution for 16 hours on an end to end rotator together with an negative control containing cellulose bandage and only cleavage buffer.
After the incubation, the supernatant containing thrombin was pink while the negative control containing only cleavage buffer was transparent with a clear pink cellulose bandage (see Figure 2). This indicates that the release mechanism work and the AsPink protein has successfully been released from the bandage into the supernatant.
Usage and Biology
This part is useful as a reporter.
asPink does not exist as CDS only but only as RBS-asPink (BBa_K1033927) and J23110-RBS-spisPink (BBa_K1033926).
iGEM2013 Uppsala: Expression of asPink in E. coli DH5alpha by promoter J23110 from medium copy plasmid pSB3K3 (left) or high copy plasmid pSB1K3 (right).
Source
The protein was first extracted and characterized by Lukyanov et. al. 2000 under the name asFP595 (GenBank: AAG02385.1). This version is codon optimized for E coli by Genscript.
References
[http://www.jbc.org/content/275/34/25879.short]Lukyanov, Konstantin A., et al. "Natural animal coloration can be determined by a nonfluorescent green fluorescent protein homolog." Journal of Biological Chemistry 275.34 (2000): 25879-25882.
[http://www.jbc.org/content/280/4/2401.short]Wilmann, Pascal G., et al. "Variations on the GFP Chromophore - A Polypeptide Fragmentation Within The Chromophore Revealed In The 2.1-Å Crystal Structure Of A Nonfluorescent Chromoprotein From Anemonia Sulcata." Journal of Biological Chemistry 280.4 (2005): 2401-2404.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]