Difference between revisions of "Ribosome Binding Sites/Prokaryotic/Constitutive/Anderson"
| Line 1: | Line 1: | ||
| − | |||
| − | |||
| − | |||
[[Image:JCA Photo.png|thumb|150px|right|The Anderson RBS family was contributed to the Registry by Prof. J. Christopher Anderson. (Photo: Peg Skorpinski)]] | [[Image:JCA Photo.png|thumb|150px|right|The Anderson RBS family was contributed to the Registry by Prof. J. Christopher Anderson. (Photo: Peg Skorpinski)]] | ||
| − | The Anderson RBS family are suitable for general protein expression in ''E. coli''. The family is known to cover a range of translation initiation rates so by testing a few family members it should be possible to find a translation initiation rate that suits your application. | + | ==Description== |
| − | + | The Anderson RBS family are suitable for general protein expression in ''E. coli''. The family is known to cover a range of translation initiation rates so by testing a few family members it should be possible to find a translation initiation rate that suits your application. The family of RBS were recovered from a library screen by [https://andersonlab.qb3.berkeley.edu/ Chris Anderson]. In the original library, 6 nucleotide locations close to where the ribosome binds were randomized (see the master sequence below). The family members have not yet been quantitatively characterized (see the Characterization section below). | |
==Obtaining Anderson RBS parts== | ==Obtaining Anderson RBS parts== | ||
| − | + | '''Via ''de novo'' synthesis''': Since the RBS parts are short sequences, they can be easily and cheaply ordered as two single-stranded complementary oligo's and annealed. See [[Ribosome Binding Sites/Construction|here]] for a tutorial on how to construct short parts via oligo annealing. | |
| − | Since the RBS are short sequences, they can be easily and cheaply ordered as two single-stranded complementary oligo's and annealed. See [[Ribosome Binding Sites/Construction|here]] for a tutorial on how to construct short parts via oligo annealing. | + | |
| − | + | '''Via the Registry distribution''': The RBS parts are included in the Registry distribution. These parts are cloned in plasmid <partinfo>pSB1A2</partinfo>, but there is also a constitutive promoter (derived from <partinfo>J23100</partinfo>) inserted into the XbaI site. So, for example, the EcoRI/PstI region of part J61100 reads:<br> | |
| − | The RBS | + | |
<font face="Courier"> | <font face="Courier"> | ||
{| style="width: 100%" align="center" | {| style="width: 100%" align="center" | ||
| Line 20: | Line 15: | ||
</font> | </font> | ||
This feature in no way prevents the use of these parts in standard Biobrick assembly. Normal prefix insertion into EcoRI/XbaI will delete this promoter element. Suffix insertion into SpeI/PstI will retain this promoter, but it can of course be removed later by a prefix insertion. Note also that the <font face="Courier" size="+1">T</font> at the start of the BioBrick suffix was randomized during library construction and is therefore rarely a <font face="Courier" size="+1">T</font>. Because the Registry does not permit variation at this position, the Registry sequence of composite parts derived from these RBS will not match the physical sequence | This feature in no way prevents the use of these parts in standard Biobrick assembly. Normal prefix insertion into EcoRI/XbaI will delete this promoter element. Suffix insertion into SpeI/PstI will retain this promoter, but it can of course be removed later by a prefix insertion. Note also that the <font face="Courier" size="+1">T</font> at the start of the BioBrick suffix was randomized during library construction and is therefore rarely a <font face="Courier" size="+1">T</font>. Because the Registry does not permit variation at this position, the Registry sequence of composite parts derived from these RBS will not match the physical sequence | ||
| + | <br style="clear:both" /> | ||
| + | ==Characterization of the Anderson RBS family== | ||
| + | [[Image:042407pic1.jpg]]<br> | ||
| + | To date, the RBS library has not been quantitatively characterized. This is largely a reflection of the method used to construct the library. Initiatially, all members of the library had the RBS variant upstream of an RFP gene. Red fluorescence for 384 individual library members, and the ranked activity of them is shown in the chart (right). All screened clones showing activity detectable above background were combined into 7 activity pools. Each pool underwent a procedure designed to remove the RFP gene and put back the SpeI site in each part. 96 individual processed clones were then sequenced, and the family of RBS parts reflects all the unique clones. | ||
| + | So, at this stage we really can't say much about their relative activity. All I can tell you is that they should all initiate translation at a rate above background, and they are very roughly in decreasing order of activity. RBS's with low numbers are likely to be stronger RBS's than those with higher numbers. | ||
| − | + | Hopefully this will change in the near future, and if that happens the info. will be posted here. | |
| + | |||
| + | ...and of course ''YOU'' could do some characterization and post the info here. | ||
| + | |||
| + | The raw data for the library after sequenceing is [[Media:030807-Bca1050 raw rbs data.xls]]. | ||
| + | The Tecan data of the clones before repooling / FokI fragmentation is [[Media:JCA_010607-TecanRBS.xls]]. | ||
| + | |||
| + | The original nomenclature was Bca1050 and then Bca9110. | ||
| + | |||
| + | |||
| + | ==Anderson RBS family members== | ||
{|class="wikitable" style="border:3px solid black; width: 60%" align="center" cellspacing="0" | {|class="wikitable" style="border:3px solid black; width: 60%" align="center" cellspacing="0" | ||
! style="background:grey; color:black" |Identifier | ! style="background:grey; color:black" |Identifier | ||
| Line 193: | Line 203: | ||
|} | |} | ||
| − | + | __NOTOC__ | |
| − | + | __NOEDITSECTION__ | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Revision as of 23:55, 6 September 2008
Description
The Anderson RBS family are suitable for general protein expression in E. coli. The family is known to cover a range of translation initiation rates so by testing a few family members it should be possible to find a translation initiation rate that suits your application. The family of RBS were recovered from a library screen by Chris Anderson. In the original library, 6 nucleotide locations close to where the ribosome binds were randomized (see the master sequence below). The family members have not yet been quantitatively characterized (see the Characterization section below).
Obtaining Anderson RBS parts
Via de novo synthesis: Since the RBS parts are short sequences, they can be easily and cheaply ordered as two single-stranded complementary oligo's and annealed. See here for a tutorial on how to construct short parts via oligo annealing.
Via the Registry distribution: The RBS parts are included in the Registry distribution. These parts are cloned in plasmid pSB1A2, but there is also a constitutive promoter (derived from BBa_J23100) inserted into the XbaI site. So, for example, the EcoRI/PstI region of part J61100 reads:
| ...Biobrick Prefix..................J23100.................XbaI....RBS Part.....Biobrick Suffix... |
| gaattcgcggccgcttctagaGTTGACGGCTAGCTCAGTCCTAGGTACAGTGCTAGCTtctagaGAAAGAGGGGACAAactagtagcggccgctgcag |
This feature in no way prevents the use of these parts in standard Biobrick assembly. Normal prefix insertion into EcoRI/XbaI will delete this promoter element. Suffix insertion into SpeI/PstI will retain this promoter, but it can of course be removed later by a prefix insertion. Note also that the T at the start of the BioBrick suffix was randomized during library construction and is therefore rarely a T. Because the Registry does not permit variation at this position, the Registry sequence of composite parts derived from these RBS will not match the physical sequence
Characterization of the Anderson RBS family
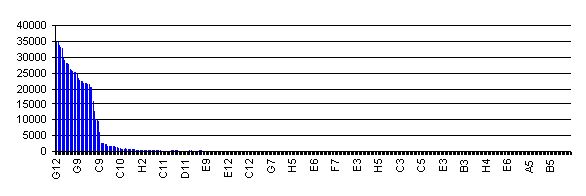
To date, the RBS library has not been quantitatively characterized. This is largely a reflection of the method used to construct the library. Initiatially, all members of the library had the RBS variant upstream of an RFP gene. Red fluorescence for 384 individual library members, and the ranked activity of them is shown in the chart (right). All screened clones showing activity detectable above background were combined into 7 activity pools. Each pool underwent a procedure designed to remove the RFP gene and put back the SpeI site in each part. 96 individual processed clones were then sequenced, and the family of RBS parts reflects all the unique clones.
So, at this stage we really can't say much about their relative activity. All I can tell you is that they should all initiate translation at a rate above background, and they are very roughly in decreasing order of activity. RBS's with low numbers are likely to be stronger RBS's than those with higher numbers.
Hopefully this will change in the near future, and if that happens the info. will be posted here.
...and of course YOU could do some characterization and post the info here.
The raw data for the library after sequenceing is Media:030807-Bca1050 raw rbs data.xls. The Tecan data of the clones before repooling / FokI fragmentation is Media:JCA_010607-TecanRBS.xls.
The original nomenclature was Bca1050 and then Bca9110.
Anderson RBS family members
| Identifier | Sequencea | Strengthb |
|---|---|---|
| Master Sequence | TCTAGAGAAAGANNNGANNNACTAGATG | |
| BBa_J61100 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61101 | TCTAGAGAAAGACAGGACCCACTAGATG | |
| BBa_J61102 | TCTAGAGAAAGATCCGATGTACTAGATG | |
| BBa_J61103 | TCTAGAGAAAGATTAGACAAACTAGATG | |
| BBa_J61104 | TCTAGAGAAAGAAGGGACAGACTAGATG | |
| BBa_J61105 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61106 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61107 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61108 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61109 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61110 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61111 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61112 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61113 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61114 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61115 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61116 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61117 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61118 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61119 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61120 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61121 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61122 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61123 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61124 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61125 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61126 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61127 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61128 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61129 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61130 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61131 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61132 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61133 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61134 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61135 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61136 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61137 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61138 | TCTAGAGAAAGAGGGGACAAACTAGATG | |
| BBa_J61139 | TCTAGAGAAAGAGGGGACAAACTAGATG |

