Difference between revisions of "Part:BBa K325909"
Jennynesje (Talk | contribs) |
Jennynesje (Talk | contribs) |
||
| Line 32: | Line 32: | ||
Team NTNU Trondheim 2017 has improved the characterisation of the biobrick BBa_K325909, which is an operon consisting of Lux C, D, A, B, E under the arabinose-induced promoter pBAD. We chose to improve this biobrick as we originally wanted to use the lux operon as a reporter for our arabinose-inducible system. In this way, we could test how the expression from the pBAD promoter was dependent on arabinose concentration. | Team NTNU Trondheim 2017 has improved the characterisation of the biobrick BBa_K325909, which is an operon consisting of Lux C, D, A, B, E under the arabinose-induced promoter pBAD. We chose to improve this biobrick as we originally wanted to use the lux operon as a reporter for our arabinose-inducible system. In this way, we could test how the expression from the pBAD promoter was dependent on arabinose concentration. | ||
<br><br> | <br><br> | ||
| − | By looking at previous characterisation results of the biobrick, we thought it would work reasonably to use DH5α competent E. coli cells and an incubation temperature of 37 °C. However, when testing with 0.01M arabinose concentrations, no observable luminescence was detected at any time after induction. Therefore, we decided to characterise the biobrick by measuring the luminescence from the lux operon at different arabinose concentrations and temperatures. We measured the luminescence with a Tecan | + | By looking at previous characterisation results of the biobrick, we thought it would work reasonably to use DH5α competent E. coli cells and an incubation temperature of 37 °C. However, when testing with 0.01M arabinose concentrations, no observable luminescence was detected at any time after induction. Therefore, we decided to characterise the biobrick by measuring the luminescence from the lux operon at different arabinose concentrations and temperatures. We measured the luminescence with a Tecan M200 plate reader, using four different temperatures and six different arabinose concentrations. As the machine could only perform heating and not cooling, it was challenging to measure at temperatures below 29 °C. |
<br><br> | <br><br> | ||
Results from our experiments are shown in the graphs below. As no luminescence was detected at 37 °C, these graphs are not included. | Results from our experiments are shown in the graphs below. As no luminescence was detected at 37 °C, these graphs are not included. | ||
Revision as of 21:47, 31 October 2017
Lux Operon (under pBAD)
Vibrio Fischeri
|
|

|
Description
This page described the lux operon from Vibrio fischeri. To relieve LuxR control we placed Lux C, D, A, B, E under the pBad promoter. A more complete description can be found on the [http://2010.igem.org/Team:Cambridge/Bioluminescence/G28 Cambridge iGEM 2010 website].
This Bacterial lux operons encodes five enzymes involved in the light-generating pathway. LuxA and LuxB encode the two subunits of the bacterial luciferase, while the products of LuxC, LuxD and LuxE synthesise the substrate for the light emitting reaction, tetradecanal. The exact function of LuxG is unknown, and it appears to be non-essential for light emission, but its presence increases light output.
In nature, the lux genes appear to be repressed by the nucleoid protein, H-NS, and occur under quorum sensing control. We removed the natural quorum sensing control to facilitate use of the part in biosensors under different regulatory inputs.
As of October 2010 we believe this is the first and only BioBrick to emit light in normal E. coli strains without the addition of any external substrate.
Measured by the [http://2010.igem.org/Team:Cambridge Cambridge iGEM team 2010]
Compatibility
Chassis: Device has been shown to work in Top 10 (Invitrogen), [http://www.ecoliwiki.net/colipedia/index.php/BW25113 BW25113 DELTA H-NS::kan] and H-NS mutant JM 230 H-NS -205::tn10.br>
Plasmids: Device has been shown to work on pSB1C3
Additional characterisation by the [http://2015.igem.org/Team:Penn Penn iGEM team 2015]
Penn iGEM 2015 used this part to make the sender cells in light-mediated cell communication. The lux operon BioBrick on PSB1C3 was transformed into NEB 10-beta competent cells (since it is a commonly used E. coli strain in iGEM) and an H-NS mutant E. coli strain (since the protein has been shown to reduce lux gene expression). The luminescent signal of the two sender strains was compared over time to determine what luminescent output trend would be best for inducing our light-sensitive receiver. Furthermore, using a calibrated conversion system designed by the team, Penn IGEM was about to convert the output from relative lights units (a measure typically used to calculate bioluminescent signals) to an absolute measure in uW/cm^2. Using the absolute units will assist in making the characterization of bioluminescent parts such as BBa_K325909 more consistent across luminometer devices and across projects. For more information, please follow this [http://2015.igem.org/Team:Penn/Sender link].
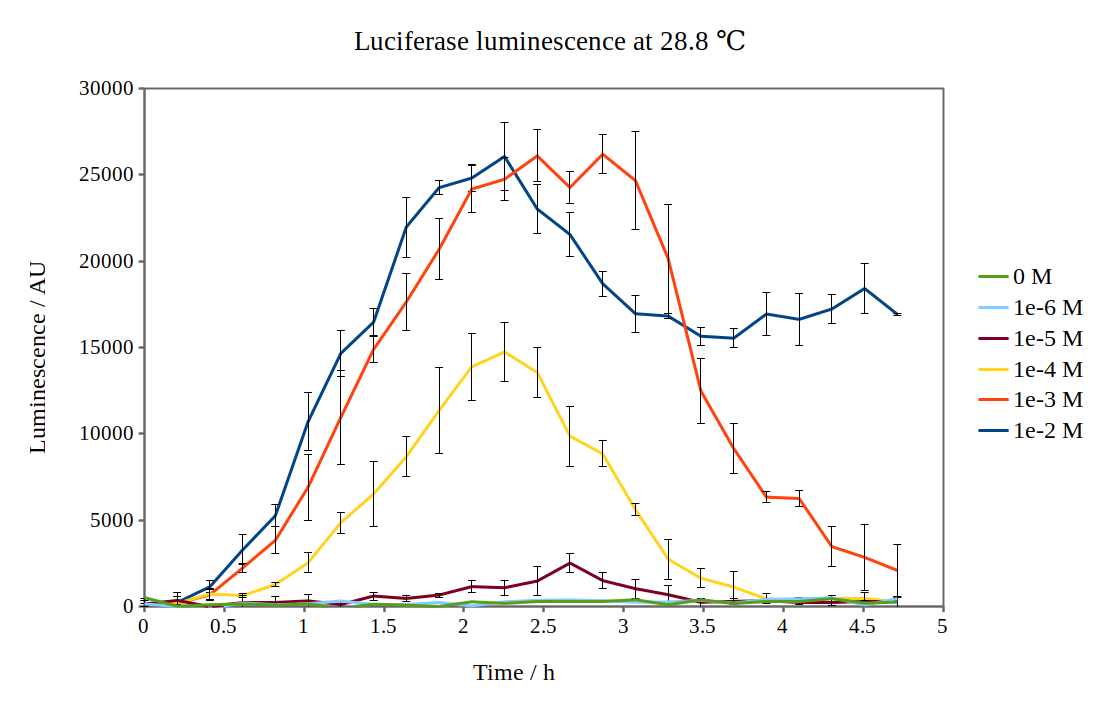
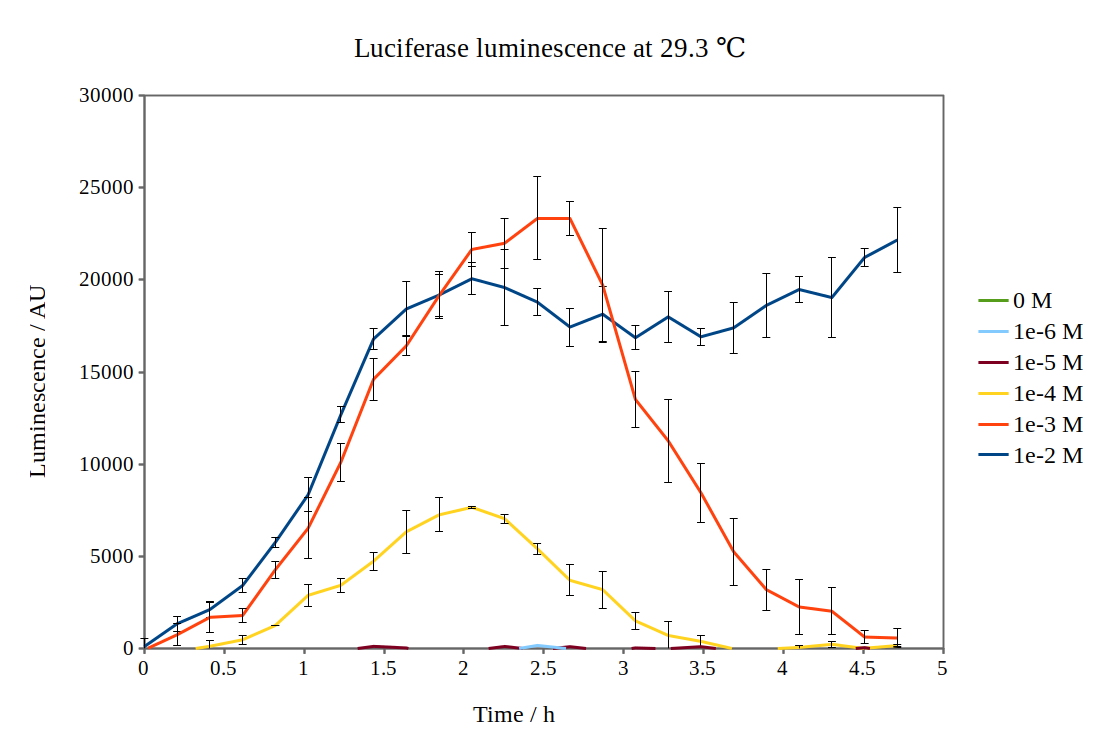
Additional characterisation by the [http://2017.igem.org/Team:NTNU_Trondheim NTNU Trondheim iGEM team 2017]
Team NTNU Trondheim 2017 has improved the characterisation of the biobrick BBa_K325909, which is an operon consisting of Lux C, D, A, B, E under the arabinose-induced promoter pBAD. We chose to improve this biobrick as we originally wanted to use the lux operon as a reporter for our arabinose-inducible system. In this way, we could test how the expression from the pBAD promoter was dependent on arabinose concentration.
By looking at previous characterisation results of the biobrick, we thought it would work reasonably to use DH5α competent E. coli cells and an incubation temperature of 37 °C. However, when testing with 0.01M arabinose concentrations, no observable luminescence was detected at any time after induction. Therefore, we decided to characterise the biobrick by measuring the luminescence from the lux operon at different arabinose concentrations and temperatures. We measured the luminescence with a Tecan M200 plate reader, using four different temperatures and six different arabinose concentrations. As the machine could only perform heating and not cooling, it was challenging to measure at temperatures below 29 °C.
Results from our experiments are shown in the graphs below. As no luminescence was detected at 37 °C, these graphs are not included.
By looking at how luciferase luminescence from the lux operon developed with time after induction by arabinose, we established that a temperature of 29 °C was optimal. This can be seen in the graph below:
Pictures
References
[http://www.jstor.org/stable/4449975 [1]:] J. Slock, (1995) Transformation Experiments Using Bioluminescence Genes of Vibrio fischeri,The American Biology Teacher, 57, 225-227.
[http://www.annualreviews.org/doi/pdf/10.1146/annurev.mi.42.100188.001055 [2]:] E.A. Meighen (1988) Enzymes and genes from the lux operons of bioluminescent bacteria, Annual Reviews in Microbiology 42, 151-176.
[http://www.annualreviews.org/doi/pdf/10.1146/annurev.ge.28.120194.001001 [3]:] E.A. Meighen, (1994) Genetics of bacterial bioluminescence, Annual Reviews of Genetics, 28, 117-139.