Difference between revisions of "Part:BBa K1033282"
| Line 20: | Line 20: | ||
<partinfo>BBa_K1033282 parameters</partinfo> | <partinfo>BBa_K1033282 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| − | ...... | + | ==Team INSA-UPS France 2017: demonstration of pGAP validity in <i>Pichia pastoris</i>== |
| + | |||
| + | We tested the functionality of pGAP as a constitutive promoter in the yeast <i>Pichia pastoris</i> background. | ||
| + | |||
| + | The biobrick was cloned in pPICZalpha vector and was integrated in <i>Pichia pastoris</i> at the pGAP genomic locus. The reporter gene here was <partinfo>BBa_K2278021</partinfo> (D-NY15 antimicrobial peptide encoding gene for our project). | ||
| + | |||
| + | |||
| + | [[Image:T--INSA-UPS_France--RTQPCRdekalitaäy.png|800px|thumb|center|<b>RTq-PCR of D-NY15 driven by BBa_K431009 promoter. </b>RNA were issued from <i>Pichia pastoris</i> strains with or without the D-NY15 encoding gene. Total RNAs were extracted from transformants and reverse transcriptions were performed using Superscript II reverse transcriptase (Invitrogen). Resulting D-NY15 cDNA were then amplified by quantitative PCR. The curves correspond to <i>P. pastoris</i> with integrated pPICZalpha-DNY15 (A1, A2, A3), water control (B1 & B2), and <i>P. pastoris</i> with integrated pPICZalpha (C1, C2, C3).]] | ||
| + | |||
| + | The amount of fluorescence provided by the qRT-PCR with the D-NY15 primers rose after 8 cycles whereas the negative control (pPICZalpha only) started to be amplified at over 29 cycles (non specific amplification). This means that the D-NY15 encoding gene is well expressed in <i>Pichia pastoris</i> with pGAP as a promoter. This promoter is therefore very efficient in the <i>Pichia pastoris</i> background. | ||
Revision as of 11:05, 27 October 2017
amilCP blue chromoprotein with strong promoter
This is a blue/purple chromoprotein (amilCP: BBa_K592009) with the RBS B0034 (BBa_B0034) and a strong promoter (CP29: BBa_K1033222). The construct has been shown to work in E. coli and should work in Lactobacillus.
(Construct name: pSB1C3-CP29-B0034-amilCP)
See http://2013.igem.org/Team:Uppsala/reporter-genes for more information.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Team INSA-UPS France 2017: demonstration of pGAP validity in Pichia pastoris
We tested the functionality of pGAP as a constitutive promoter in the yeast Pichia pastoris background.
The biobrick was cloned in pPICZalpha vector and was integrated in Pichia pastoris at the pGAP genomic locus. The reporter gene here was BBa_K2278021 (D-NY15 antimicrobial peptide encoding gene for our project).
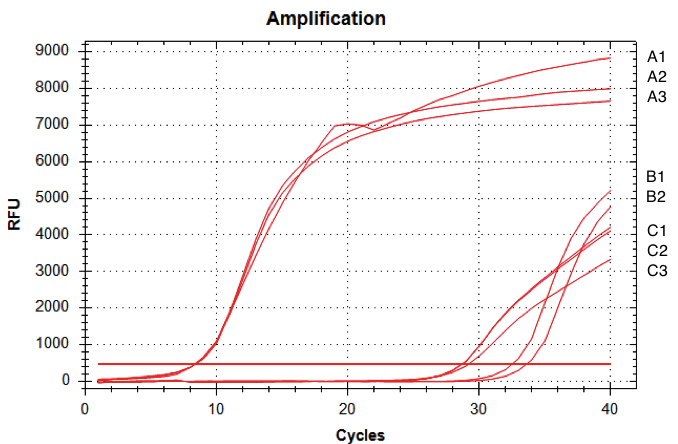
The amount of fluorescence provided by the qRT-PCR with the D-NY15 primers rose after 8 cycles whereas the negative control (pPICZalpha only) started to be amplified at over 29 cycles (non specific amplification). This means that the D-NY15 encoding gene is well expressed in Pichia pastoris with pGAP as a promoter. This promoter is therefore very efficient in the Pichia pastoris background.
