Difference between revisions of "Part:BBa K1976048"
(→Characteristics) |
|||
| Line 79: | Line 79: | ||
<partinfo>BBA_K1976048 SequenceAndFeatures</partinfo> | <partinfo>BBA_K1976048 SequenceAndFeatures</partinfo> | ||
<br> | <br> | ||
| + | ===Characterization from Igem Team UNILausanne 2020=== | ||
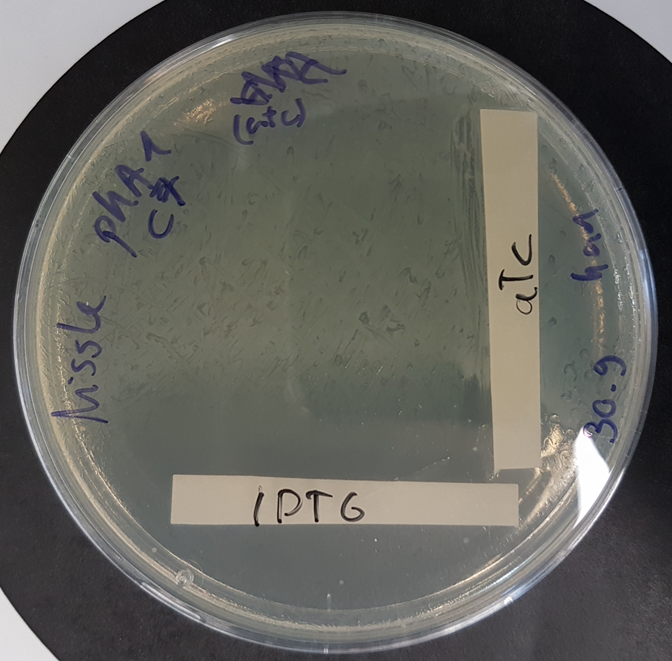
| + | Functional kill switch assay with IPTG and aTc gradients on agar plate | ||
| + | |||
| + | [[File:BBa K3482016 kill-switch plate.png|200px|]] | ||
| + | |||
| + | E.coli Nissle 1917 were transformed with a plasmid containing the part BBa K3482016 and the part <html><a style="padding: 0px; margin: 0px;" href="https://parts.igem.org/Part:BBa_K3482020"> BBa_K3482020 </a></html> (CcdA antitoxin part) and plated with a gradient of aTc and IPTG on agar plate. | ||
| + | The plate shows strong activity of the CcdA antitoxin with aTc induction, whereas IPTG induction promotes production of the CcdB toxin, resulting in a number of surviving cells (probable mutants) proportional to the dilution of the plated culture. | ||
| + | |||
| + | |||
| + | |||
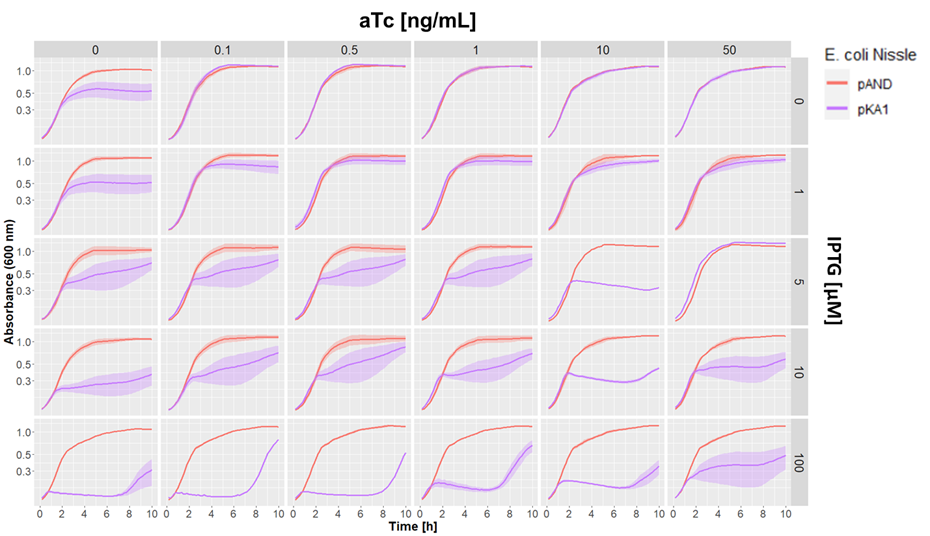
| + | We aslo tested our pKA1 plasmid encoding for the ccdB toxin and ccdA antitoxin in <i>E. coli Nissle 1917 ΔclbA</i>. We used different concentrations of IPTG and aTc to study the effect of differential expression of the toxin and antitoxin on the growth of our strain. We compared this to the strain transformed with the empty vector pAND | ||
| + | |||
| + | [[File:BBa K3482004 atciptg.png|800px]] | ||
| + | |||
| + | Dose-response growth curve of <i>E. coli Nissle 1917 ΔclbA</i> harboring kill switch plasmid pKA1 at 37°C. <i>E. coli Nissle</i> with pAND (red line), <i>E. coli Nissle</i> with pKA1 (blue line). The lines and shade represent the mean ± standard error. | ||
| + | |||
| + | We observed desired growth inhibition of the strain with pKA1 at high IPTG and low aTc concentrations, while the pAND strain showed no alteration in growth in any of the tested conditions. The uniformity of these growth curves of the strain with pAND demonstrates that the standardized culturing and OD normalization of our protocol can yield highly reproducible results. | ||
| + | |||
| + | We saw an effect of IPTG on growth inhibition starting from 5 µM IPTG. At maximum IPTG concentration, we observed complete absence of growth of the pKA1 strain for the first 7 h. However, afterwards, we observed that the bacteria were able to grow. This was probably due to mutations in the kill switch system. In contrast, at lower IPTG concentrations (5 and 10 µM), we still observed an inhibitor effect on growth, which albeit being less strong, was more constant and persisted for at least 10 h. This suggests that decreasing the expression of the toxin lowers the selective pressure on the kill switch system, which makes this configuration more evolutionarily stable. In fact, while we observe a strong uninhibited growth of escape mutants after 7 h with 100 µM IPTG, at 10 µM IPTG the cells show a constant repression on their growth rate, which indicates that no or little escape mutants have emerged. | ||
| + | |||
| + | Overall, we demonstrated that the ccdB/ccdA encoding pKA1 plasmid could efficiently inhibit the growth of <i>E. coli Nissle 1917</i> at 100 µM IPTG for up to 7 h, and provided even longer growth inhibition at 5 and 10 µM IPTG. | ||
<br> | <br> | ||
Revision as of 00:26, 28 October 2020
Colicin E2 DNase domain "miniColicin"
Colicin E2 is a peptide, that is toxic to coliform bacteria. It is one of a group of proteins of this type, it being a nicking endonuclease. It has a corresponding activity inhibiting protein called immunity protein Im2, which is also in the registry under the name BBa_K1976027.
Figure 1:Structure of Colicin E2 DNase domain derived from a molecular dynamics simulation.
Contents
Usage
The Colicin E2 immunity protein (BBa_K1976027 and BBa_K1976028) can be used in combination with the Colicin E2 DNase. Due to to the high binding affinity of the immunity protein to the DNase domain (Kd ∼ 10− 15 M)2, it can be used to fuse proteins and in combination with a suppression mechanism for the immunity protein as a bacterial killswitch.
Characteristics
| Molecular Weight | 15.2 kDa |
| Residues | 133 |
| pI | 9.06 |
| [Data taken created in Snapgene and http://isoelectric.ovh.org/calculate.php] |
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 28
- 1000COMPATIBLE WITH RFC[1000]
Characterization from Igem Team UNILausanne 2020
Functional kill switch assay with IPTG and aTc gradients on agar plate
E.coli Nissle 1917 were transformed with a plasmid containing the part BBa K3482016 and the part BBa_K3482020 (CcdA antitoxin part) and plated with a gradient of aTc and IPTG on agar plate. The plate shows strong activity of the CcdA antitoxin with aTc induction, whereas IPTG induction promotes production of the CcdB toxin, resulting in a number of surviving cells (probable mutants) proportional to the dilution of the plated culture.
We aslo tested our pKA1 plasmid encoding for the ccdB toxin and ccdA antitoxin in E. coli Nissle 1917 ΔclbA. We used different concentrations of IPTG and aTc to study the effect of differential expression of the toxin and antitoxin on the growth of our strain. We compared this to the strain transformed with the empty vector pAND
Dose-response growth curve of E. coli Nissle 1917 ΔclbA harboring kill switch plasmid pKA1 at 37°C. E. coli Nissle with pAND (red line), E. coli Nissle with pKA1 (blue line). The lines and shade represent the mean ± standard error.
We observed desired growth inhibition of the strain with pKA1 at high IPTG and low aTc concentrations, while the pAND strain showed no alteration in growth in any of the tested conditions. The uniformity of these growth curves of the strain with pAND demonstrates that the standardized culturing and OD normalization of our protocol can yield highly reproducible results.
We saw an effect of IPTG on growth inhibition starting from 5 µM IPTG. At maximum IPTG concentration, we observed complete absence of growth of the pKA1 strain for the first 7 h. However, afterwards, we observed that the bacteria were able to grow. This was probably due to mutations in the kill switch system. In contrast, at lower IPTG concentrations (5 and 10 µM), we still observed an inhibitor effect on growth, which albeit being less strong, was more constant and persisted for at least 10 h. This suggests that decreasing the expression of the toxin lowers the selective pressure on the kill switch system, which makes this configuration more evolutionarily stable. In fact, while we observe a strong uninhibited growth of escape mutants after 7 h with 100 µM IPTG, at 10 µM IPTG the cells show a constant repression on their growth rate, which indicates that no or little escape mutants have emerged.
Overall, we demonstrated that the ccdB/ccdA encoding pKA1 plasmid could efficiently inhibit the growth of E. coli Nissle 1917 at 100 µM IPTG for up to 7 h, and provided even longer growth inhibition at 5 and 10 µM IPTG.
[1] Cascales et al, Colicin Biology, Microbiology and Molecular Biology Reviews, vol. 71, pp. 158-229, 2007
[2] Wojdyla et al, Journal of Molecular Biology, vol. 417, Issues 1–2, 16 March 2012, Pages 79–94