Difference between revisions of "Part:BBa K1949101"
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1949101 short</partinfo> | <partinfo>BBa_K1949101 short</partinfo> | ||
| − | <span style="margin-left: 10px;">We designed parts(<partinfo>BBa_1949100</partinfo>, <partinfo>BBa_1949101</partinfo>, <partinfo>BBa_1949102</partinfo>, <partinfo>BBa_1949103</partinfo>, <partinfo>BBa_1949104</partinfo>). | + | <span style="margin-left: 10px;">We designed parts(<partinfo>BBa_1949100</partinfo>), <partinfo>BBa_1949101</partinfo>, <partinfo>BBa_1949102</partinfo>, <partinfo>BBa_1949103</partinfo>, <partinfo>BBa_1949104</partinfo>). |
===Characterize === | ===Characterize === | ||
Revision as of 12:54, 19 October 2016
PBAD-rbs-mazF
We designed parts(No part name specified with partinfo tag.), No part name specified with partinfo tag., No part name specified with partinfo tag., No part name specified with partinfo tag., No part name specified with partinfo tag.).
Characterize
We performed experiment to work our final genetic circuits and characterized mazE and mazF.
This experiment consists of the four parts below.
Ⅰ.Adjustment of MazF Expression
Ⅱ.mazEF System Assay ~Stop & GO~
Ⅲ.mazEF System Assay ~Go & Stop~
Ⅳ.Control of Cell Growth
Ⅰ.Adjustment of MazF Expression
In order to control the TA system well, it is necessary to adjust the expression of MazF as a toxin. It has been reported that MazF has very strong ability to inhibit cell growth, and antitoxins cannot recover it under overexpression of MazF. Therefore, we explored the conditions that can control the TA system by adjusting the expression of MazF.
We constructed
GFP : Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3)
MazF : PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3).
result
It was found that the cell growth of E. coli B was inhibited when the concentration of arabinose, the inducer of MazF, is more than 0.02% (Fig. 1.). However, when arabinose concentration was 0.2%, GFP fluorescence of both of E. coli intensity fell markedly.
Ⅱ.mazEF System Assay ~Stop & GO~
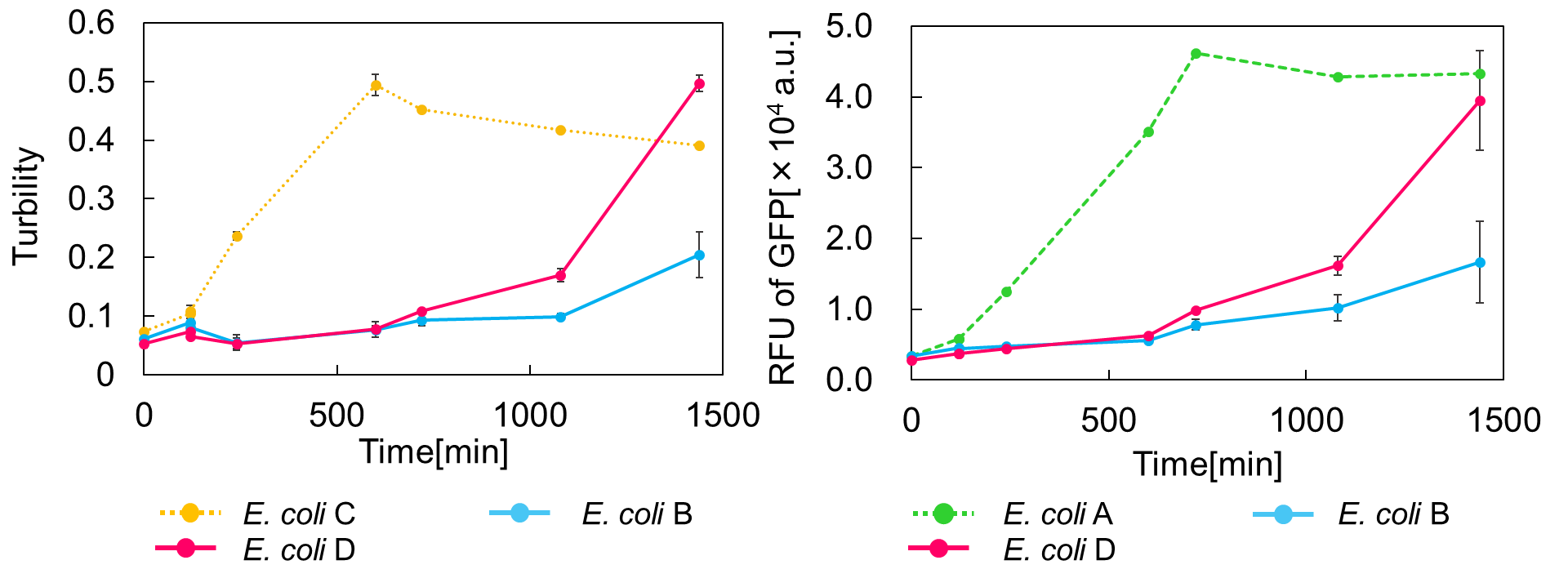
The biggest attraction of the TA system is that it is able to control cell growth and synthesis of protein. In this experiment, MazE expression was induced by the addition of IPTG(2 mM) after MazF expression was induced by the addition of arabinose(0.02%). As a result, it was able to resuscitate from a state of being inhibited cell growth. We named this experiment as "Stop & Go" because it was to resuscitate growth from inhibiting cell growth.
We constructed
vector : PBAD - rbs (pSB6A1), Plac - rbs (pSB3K3)
GFP : Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3)
MazF + MazE : PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Plac - rbs - mazE (pSB3K3)
MazF : PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3).
result
It was found from and that MazF inhibited cell growth(Fig. 2.). MazE was induced 2 h after mazE expression, and about 8 h later, cell growth was recovered that had stopped.
From these results, it was suggested that E. coli whose cell growth was inhibited by MazF was able to resuscitate by expression of mazE.
MazF is induced by the addition of arabinose, MazE induced by the addition of IPTG. At time zero, mazF expression was induced by the addition of 0.02% arabinose to the culture.
Ⅲ.mazEF System Assay ~Go & Stop~
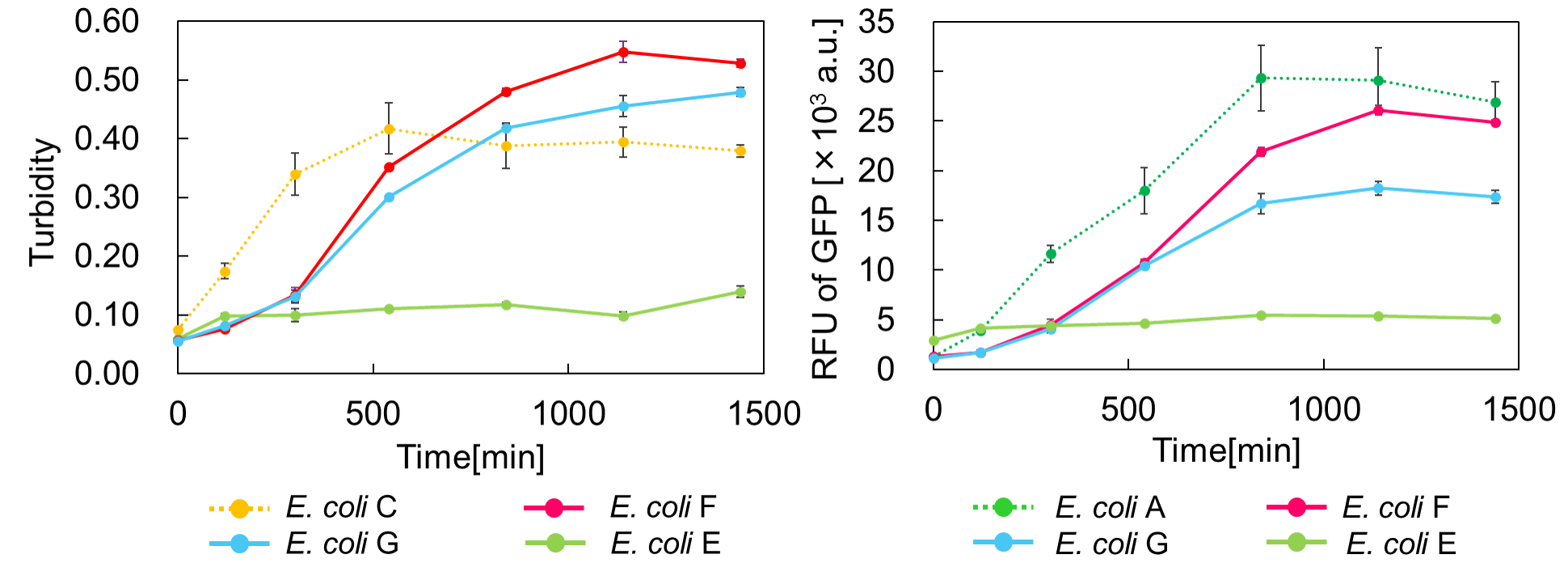
We found that a toxin inhibits cell growth, and an antitoxin resuscitates it. However, what will happen when a toxin is expressed after the antitoxin constitutive expression? Therefore, we conducted the experiment. Since cell growth was resuscitated after cells had grown, we named this experiment, "Go & Stop".
We constructed
vector : PBAD - rbs(pSB6A1), Plac - rbs (pSB3K3)
GFP : Pcon - rbs - gfp (pSB6A1), Plac - rbs(pSB3K3)
MazF + MazE(weak) : PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Pcon - rbs(weak) - mazE (pSB3K3)
MazF + MazE : PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Pcon - rbs - mazE (pSB3K3)
MazF : PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), vector (pSB3K3).
result
E. coli encoded mazE which is on downstream of weak RBS(BBa_J61117) has more turbidity than E. coli encoded mazE which is on downstream of normal RBS(BBa_B0034)(Fig. 3.).
Both of those E. coli would reach Stationary phase when there is little RFU of GFP. E. coli encoded mazE which is on downstream of normal RBS reached almost the same stationary phase as E. coli without TA system.
MazF induced by the addition of arabinose, MazE induced by the addition of IPTG. At time zero, mazF expression was induced by the addition of 0.02% arabinose to the culture.
Ⅳ.Control of Cell Growth
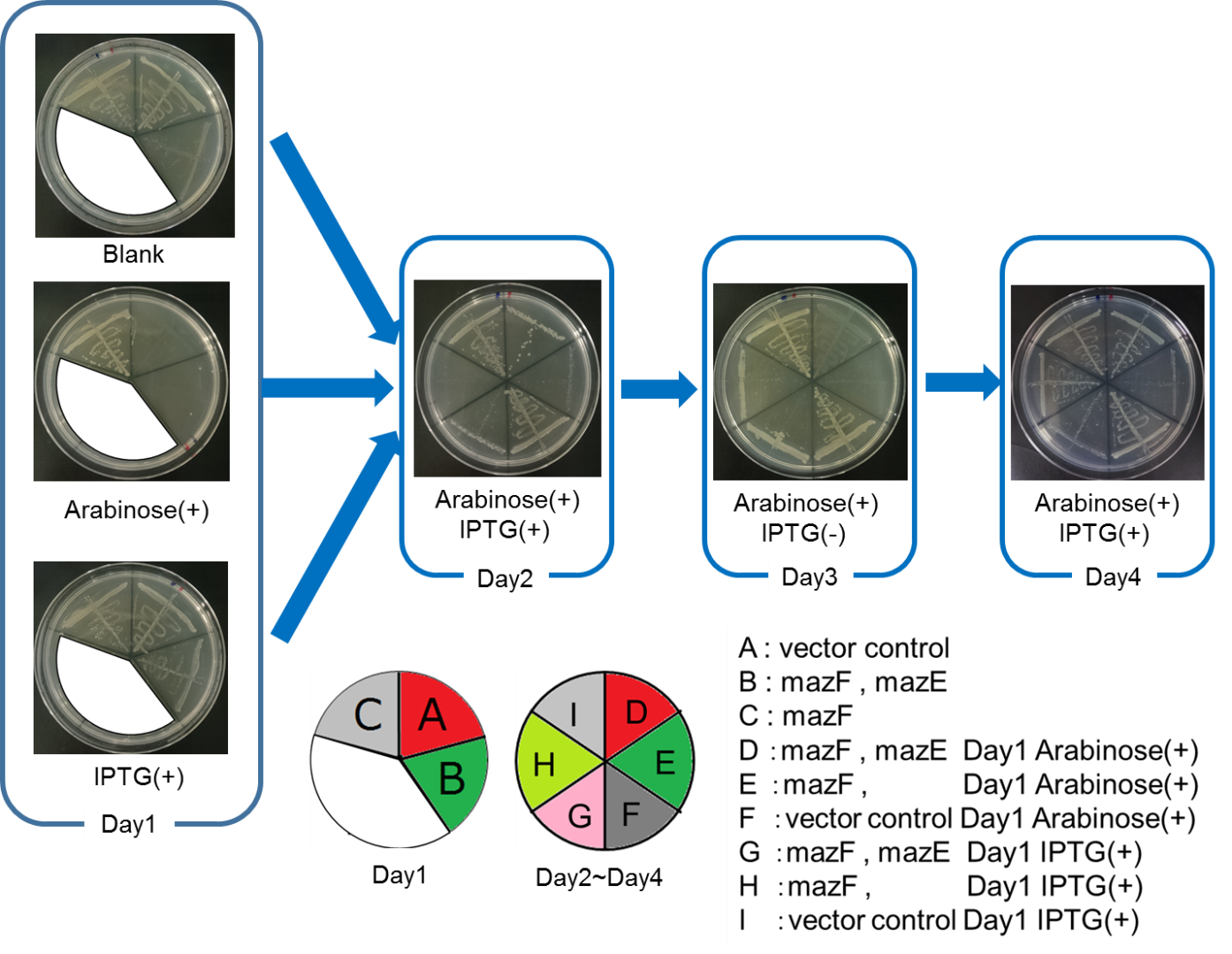
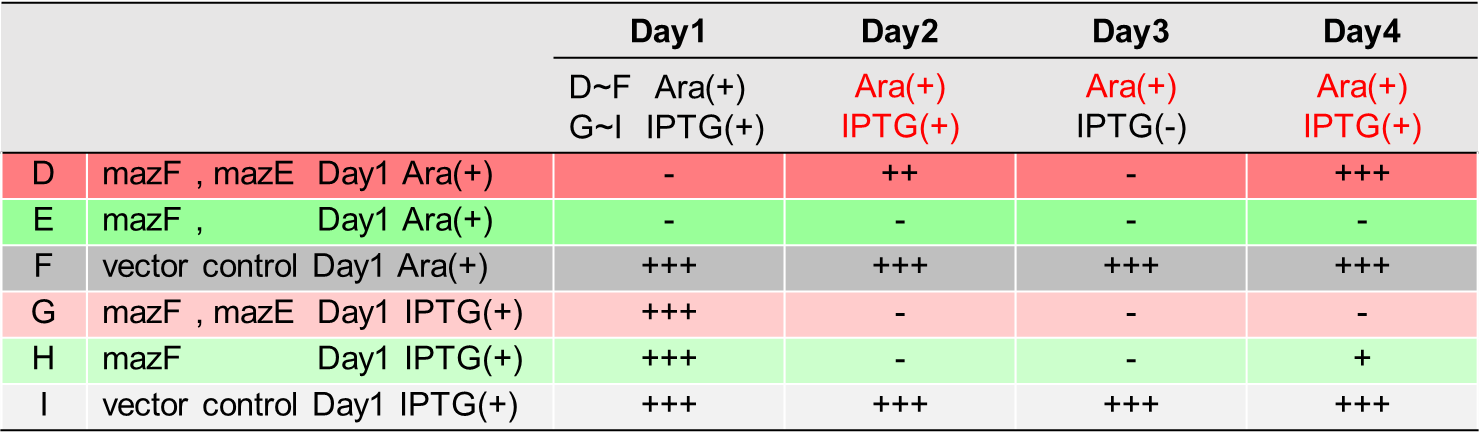
The control of cell growth by the mazEF system has been shown until the previous sections. In this section, we analyzed whether the “stop & go” experiment can be repeated many times.
We constructed
vector : PBAD - rbs (pSB6A1), Plac - rbs (pSB3K3)
MazF + MazE : PBAD - rbs - mazF (pSB6A1), Plac - rbs - mazE (pSB3K3)
MazF : PBAD - rbs - mazF(pSB6A1), Plac - rbs (pSB3K3).
result
From the result(Fig. 4., Fig. 5.), it was clarified that growth of E. coli cells was repeatedly controlled by expression of maze. The results of this experiment are very useful in our project, and it is expected to lead to new biotechnological applications.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1205
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1144
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 979
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 961