Difference between revisions of "Part:BBa K823052"
| Line 7: | Line 7: | ||
This BioBrick was used for promoter evaluation in ''B. subtilis''. | This BioBrick was used for promoter evaluation in ''B. subtilis''. | ||
| + | |||
| + | Find out more about the design of our [https://static.igem.org/mediawiki/parts/b/b9/LMU-Munich_2012_Our_Freiburg_standard_FusionPrefix.pdf prefix with ribosome binding site]. | ||
prefix: GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC | prefix: GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC | ||
| Line 13: | Line 15: | ||
| − | + | We used this ''lacZ'' gene to evaluate our promoters [https://parts.igem.org/Part:BBa_K823000 PliaG], [https://parts.igem.org/Part:BBa_K823001 PliaI], [https://parts.igem.org/Part:BBa_K823003 Pveg]. | |
| − | + | ||
| − | We used this ''lacZ'' gene to evaluate our promoters. | + | |
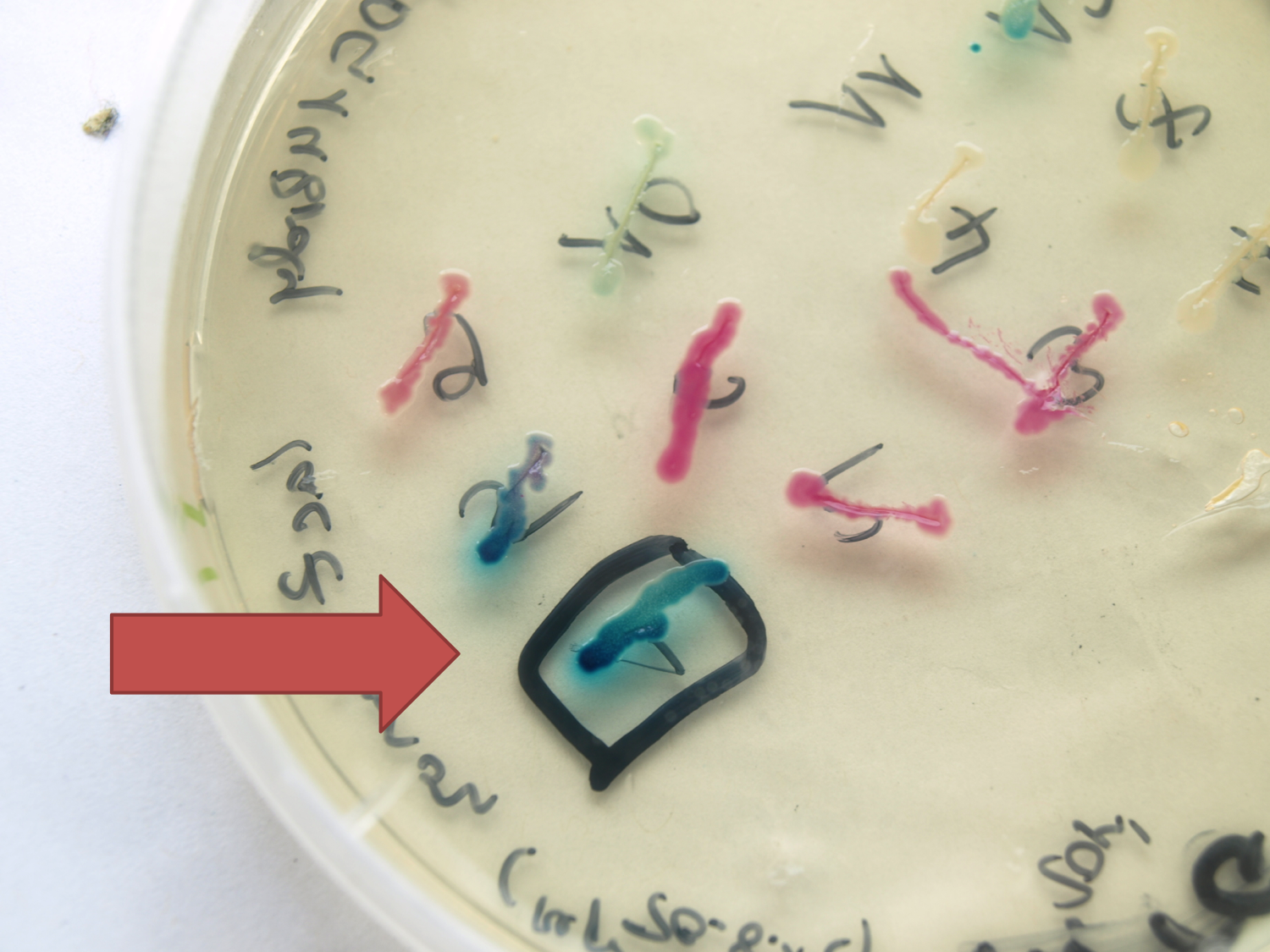
[[Image:LacZ plate.png|thumb|center|400px|lacZ under the control of Pspac in pSB1C3. Plate was induced with xylose.]] | [[Image:LacZ plate.png|thumb|center|400px|lacZ under the control of Pspac in pSB1C3. Plate was induced with xylose.]] | ||
Revision as of 08:59, 26 September 2012
lacZ, B. subtilis optimized + RBS + Term
lacZ in Freiburg standard, Bacillus subtilis codon optimized with ribosome binding site and terminator (B0014).
lacZ is a β-galactosidase which cleaves the substrates X-Gal (5-bromo-4-chloro-indolyl-β-D-galactopyranoside) and ONPG (ortho-Nitrophenyl-β-galactoside).
This BioBrick was used for promoter evaluation in B. subtilis.
Find out more about the design of our prefix with ribosome binding site.
prefix: GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC
suffix: ACCGGTTAATACTAGTAGCGGCCGCTGCAGT
We used this lacZ gene to evaluate our promoters PliaG, PliaI, Pveg.
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]