Part:BBa_K823000
PliaG
PliaG is the promoter of the liaG gene of Bacillus subtilis, which is weak and constitutive.
Usage and Biology
PliaG is a weak, constitutive promoter from B. subtilis. It is responsible for the transcription of the last four genes of the liaIHGFSR locus and therefore for the production of the components of the LiaRS system, which is important for the detection of cell wall antibiotics [http://www.ncbi.nlm.nih.gov/pubmed?term=Journal%20of%20Bacteriology%2C%20188%20%2814%29%3A%205153%E2%80%935166: (Jordan et al., 2006)]. PliaG was evaluated with the lux operon as well as the lacZ as reporter. For more details visit the [http://2012.igem.org/Team:LMU-Munich/Data/Constitutive Data] page of the LMU-Munich Team 2012 or get an overview of our whole project [http://2012.igem.org/Team:LMU-Munich Beadzillus].
Evaluation
This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis] by Radeck et al..
Luminescence measurements
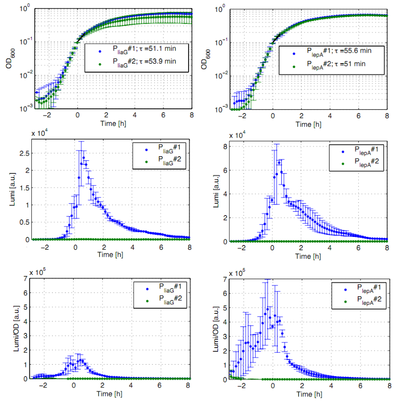
The constitutive promoters PliaG and PlepA were evaluated in the reporter vector pSBBs3C-luxABCDE which contains the lux operon. The promoter activity leads to gene expression and to the production of the protein luciferase. The luminescence produced by this protein can be measured with the plate reader Synergy2 (Biotek) (Fig.1).
|
All clones show a normal growth behaviour. The activity of both promoters increases during transition from log to stationary phase. PliaG has an activity maximum of about 100.000 Lumi/OD600. PlepA shows a maximum of about 400.000 Lumi/OD600. Comparing these two constitutive promoters the activity of PlepA is about four times higher than the activity of PliaG. In the late stationary phase the activity completely disappears. The second clone of the promoters PlepA and PliaG did not show any luminescence activity. Therefore additional clones should be measured.
β-galactosidase assay
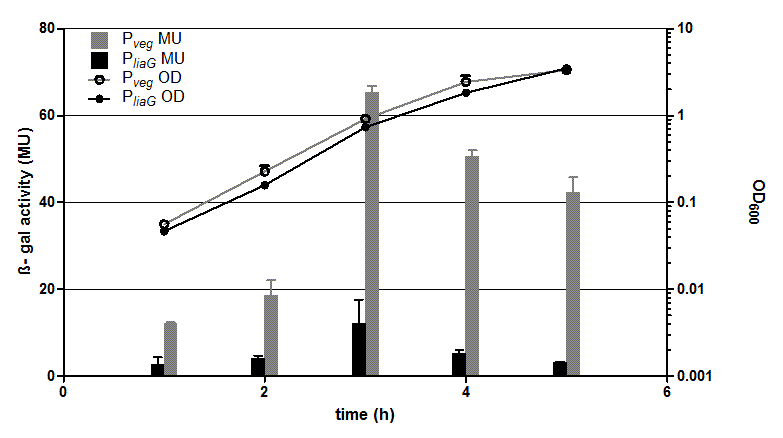
The two constitutive promoters PliaG and Pveg were evaluated with the reporter vector pSBBs1C-lacZ which contains the lacZ reporter gene (Fig.2).
|
Promoter activity leads to the expression of the β-galactosidase. The β-galactosidase assay of the constitutive Bacillus promoters Pveg and PliaG was repeated three times. The graph shows data of one representative experiment. In the beginning of the growth curve both promoters show only low activity before it increases to a maximum before it decreases to the begininng level after about seven hours (Data not shown). Summing up, the course of activity of both promoters Pveg and PliaG is very similar based on the growth curve. The highest β-galactosidase activity and therefore the highest activity of the promoter Pveg with a maximum of 65 Miller units can be found during the transition from the logarithmic to the stationary phase. This is about five times higher than the acitivity of the promoter PliaG with a maximum activity of about 12 Miller Units.
Sequence and Features
This part was amplified from the genome of B. subtilis.
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//promoter
//regulation/constitutive
//rnap/prokaryote/subtilis/sigmaa
| None |