Difference between revisions of "Template:HelpPage/Assembly Methods"
m |
m |
||
| Line 2: | Line 2: | ||
===3A Assembly=== | ===3A Assembly=== | ||
| − | [[Image:3AAssembly.png|thumb|right| | + | [[Image:3AAssembly.png|thumb|right|250px|Image: To be replaced]] |
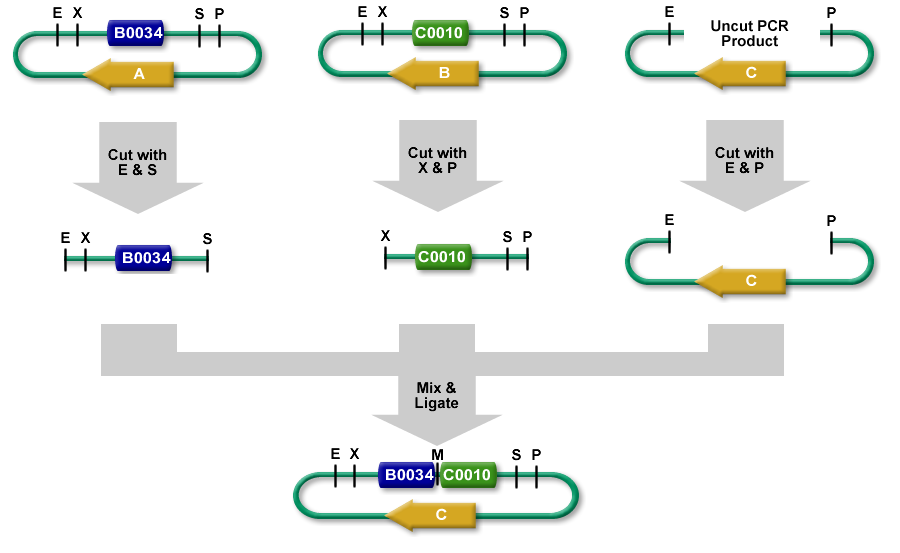
| − | 3A Assembly (which stands for three antibiotic assembly) is a method for assembling two | + | 3A Assembly (which stands for three antibiotic assembly) is a method for assembling two part samples and selecting for correct assemblies through antibiotics. 3A assembly uses the restriction sites on the prefix and suffix to assemble part samples. This new composite part will maintain the same prefix and suffix as its "parents" and contain a scar, where the cut and re-ligated restriction sites were stitched together. |
| − | It uses effective antibiotic selection to eliminate unwanted background colonies and eliminates the need for gel purification and colony PCR of the resulting colonies. In theory, about 97% of the colonies should be the desired assembly. | + | It uses effective antibiotic selection to eliminate unwanted background colonies and eliminates the need for gel purification and colony PCR of the resulting colonies. In theory, about 97% of the colonies should be the desired assembly. Additionally, the Registry has been providing iGEM teams and Registry labs with linearized plasmid backbones to further improve this assembly method. |
| − | + | ====3A Assembly Advantages==== | |
| − | + | ||
| − | + | ||
| − | + | ||
*No PCR | *No PCR | ||
| − | *No | + | *No gel purification |
| − | * | + | *Higher success rate compared to Standard Assembly. |
| − | + | ====Links==== | |
| − | + | *[[Help:Assembly/3A_Assembly|3A Assembly Overview]] | |
| − | + | *[[Help:Protocol/3A_Assembly|3A Assembly Protocol]] | |
| − | [[Help:Protocol/3A_Assembly|3A | + | |
===Scarless Assembly=== | ===Scarless Assembly=== | ||
| − | There are assembly methods which also allow for the construction of composite parts without scars or specific linkers. Additionally, these methods may be able to circumvent assembly standard incompatibility between part samples: a part sample in a Silver RFC[23] plasmid backbone can be assembled with a part sample in a Berkeley RFC[21] plasmid backbone. | + | There are assembly methods which also allow for the construction of composite parts without scars or specific linkers, which is particularly useful for the assembly of proteins. Additionally, these methods may be able to circumvent assembly standard incompatibility between part samples: a part sample in a Silver RFC[23] plasmid backbone can be assembled with a part sample in a Berkeley RFC[21] plasmid backbone. |
| − | ==== | + | ====Gibson Assembly==== |
| + | Gibson Assembly has not been tested by the Registry yet, but several teams have had success with this assembly method. The [http://2010.igem.org/Team:Cambridge/Gibson/Introduction Cambridge 2010 iGEM Team] developed a set of protocols and tools that may be useful. | ||
| + | |||
| + | The Registry will be evaluating Gibson Assembly, and will have resources available for this assembly method available soon. | ||
| + | |||
| + | ====DNA Synthesis==== | ||
| + | Just as DNA synthesis can be used to construct a new basic part, it may also be used to assemble parts together. Using the Registry database you can pull the sequence for parts you're interested, put them together in series, and then send the new composite sequence off to be synthesized. | ||
===Standard Assembly=== | ===Standard Assembly=== | ||
| − | [[Image:StdAss.png|thumb|right| | + | [[Image:StdAss.png|thumb|right|250px|Image: To be replaced]] |
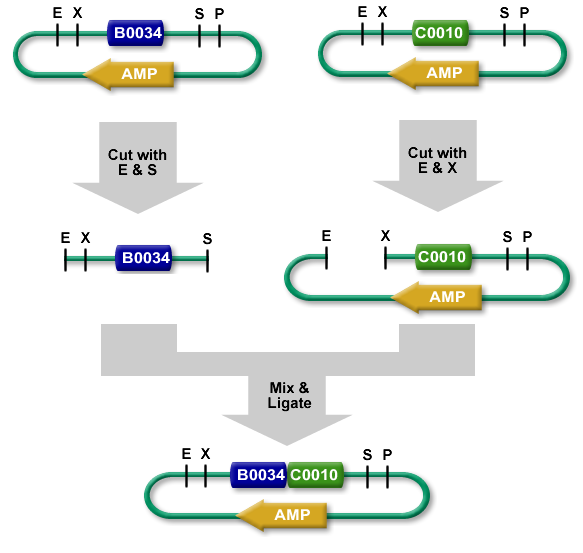
| − | For Standard | + | For Standard Assembly, a part sample is cut out from its plasmid backbone and inserted into the prefix of a plasmid backbone of another part. Two restriction digests are done, one for the part sample that will be moved and one for the plasmid backbone that will receive it. The digests are then run on a gel and using gel purification the required fragments are isolated (the part sample and the cut plasmid backbone). The purified insert and cut plasmid backbone are ligated and the resulting composite part can be transformed into E.coli cells. |
| + | |||
| + | Due to its use of gel purification and lower success rate, the Registry no longer recommends the Standard Assembly method. | ||
| − | ==== | + | ====Links==== |
| + | *[[Help:Assembly/Standard_Assembly|Standard Assembly Overview]] | ||
| + | *[[Help:Protocol/Standard_Assembly|Standard Assembly Protocol]] | ||
Revision as of 16:55, 2 March 2012
There are different methods for assembling parts: some are due to different standards requiring different materials and methods (i.e. restriction enzymes), others are due to preferences in protocol (faster assembly, higher efficiency, ease of use, etc).
Contents
3A Assembly
3A Assembly (which stands for three antibiotic assembly) is a method for assembling two part samples and selecting for correct assemblies through antibiotics. 3A assembly uses the restriction sites on the prefix and suffix to assemble part samples. This new composite part will maintain the same prefix and suffix as its "parents" and contain a scar, where the cut and re-ligated restriction sites were stitched together.
It uses effective antibiotic selection to eliminate unwanted background colonies and eliminates the need for gel purification and colony PCR of the resulting colonies. In theory, about 97% of the colonies should be the desired assembly. Additionally, the Registry has been providing iGEM teams and Registry labs with linearized plasmid backbones to further improve this assembly method.
3A Assembly Advantages
- No PCR
- No gel purification
- Higher success rate compared to Standard Assembly.
Links
Scarless Assembly
There are assembly methods which also allow for the construction of composite parts without scars or specific linkers, which is particularly useful for the assembly of proteins. Additionally, these methods may be able to circumvent assembly standard incompatibility between part samples: a part sample in a Silver RFC[23] plasmid backbone can be assembled with a part sample in a Berkeley RFC[21] plasmid backbone.
Gibson Assembly
Gibson Assembly has not been tested by the Registry yet, but several teams have had success with this assembly method. The [http://2010.igem.org/Team:Cambridge/Gibson/Introduction Cambridge 2010 iGEM Team] developed a set of protocols and tools that may be useful.
The Registry will be evaluating Gibson Assembly, and will have resources available for this assembly method available soon.
DNA Synthesis
Just as DNA synthesis can be used to construct a new basic part, it may also be used to assemble parts together. Using the Registry database you can pull the sequence for parts you're interested, put them together in series, and then send the new composite sequence off to be synthesized.
Standard Assembly
For Standard Assembly, a part sample is cut out from its plasmid backbone and inserted into the prefix of a plasmid backbone of another part. Two restriction digests are done, one for the part sample that will be moved and one for the plasmid backbone that will receive it. The digests are then run on a gel and using gel purification the required fragments are isolated (the part sample and the cut plasmid backbone). The purified insert and cut plasmid backbone are ligated and the resulting composite part can be transformed into E.coli cells.
Due to its use of gel purification and lower success rate, the Registry no longer recommends the Standard Assembly method.