Difference between revisions of "Part:BBa K584001"
| Line 4: | Line 4: | ||
This part was designed to test the functionality of the constitutive promotor region. | This part was designed to test the functionality of the constitutive promotor region. | ||
| − | Characterization | + | <b>Characterization</b> |
<p align="justify">We fused the constitutive promoter to a GFP reporter, and assayed the promoter’s activity after addition of different amounts of arabinose and IPTG. As such these results can serve as a control for the results obtained for the arabinose-inducible [https://parts.igem.org/wiki/index.php?title=Part:BBa_K584000 BBa_K584000] and IPTG-inducible [https://parts.igem.org/wiki/index.php?title=Part:BBa_K584002 BBa_K584002] bricks. | <p align="justify">We fused the constitutive promoter to a GFP reporter, and assayed the promoter’s activity after addition of different amounts of arabinose and IPTG. As such these results can serve as a control for the results obtained for the arabinose-inducible [https://parts.igem.org/wiki/index.php?title=Part:BBa_K584000 BBa_K584000] and IPTG-inducible [https://parts.igem.org/wiki/index.php?title=Part:BBa_K584002 BBa_K584002] bricks. | ||
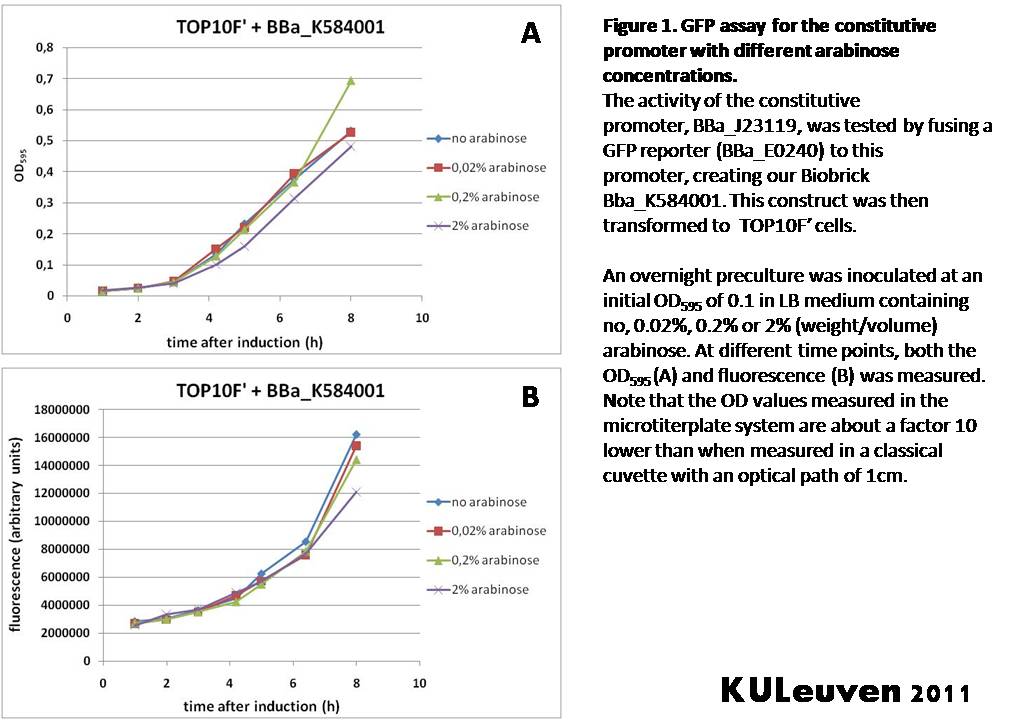
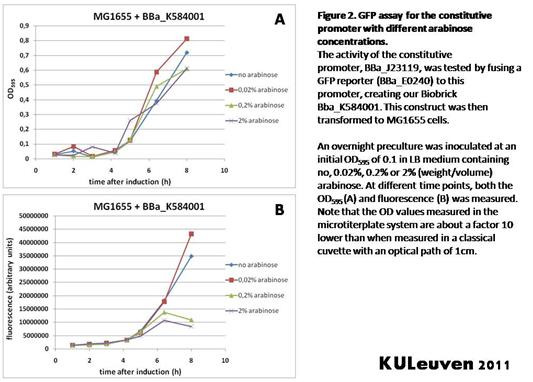
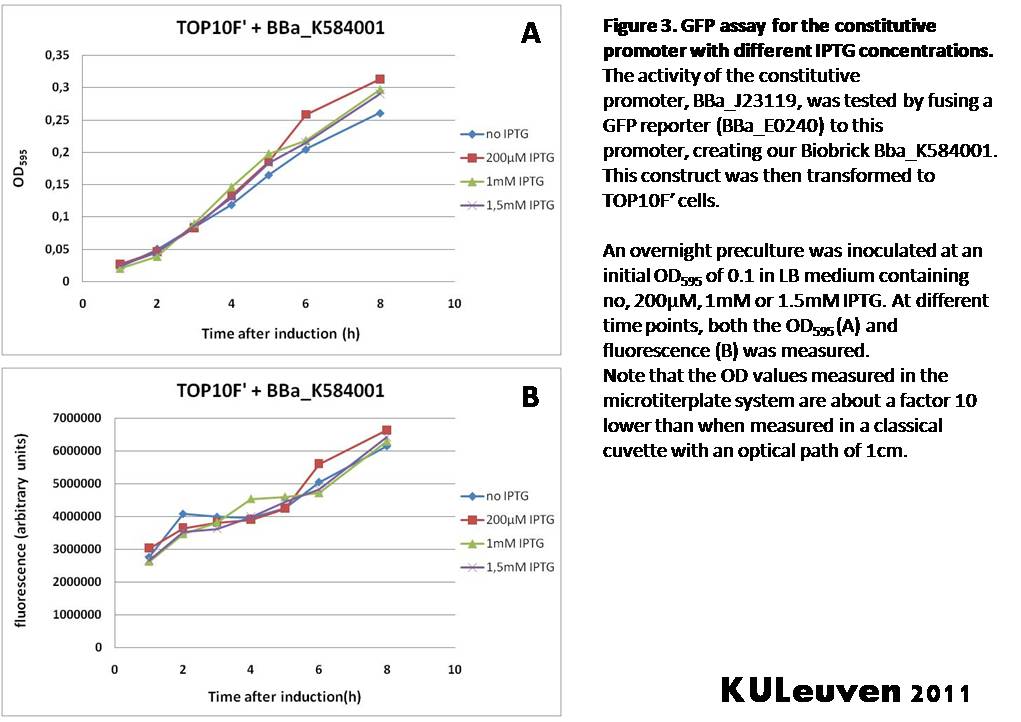
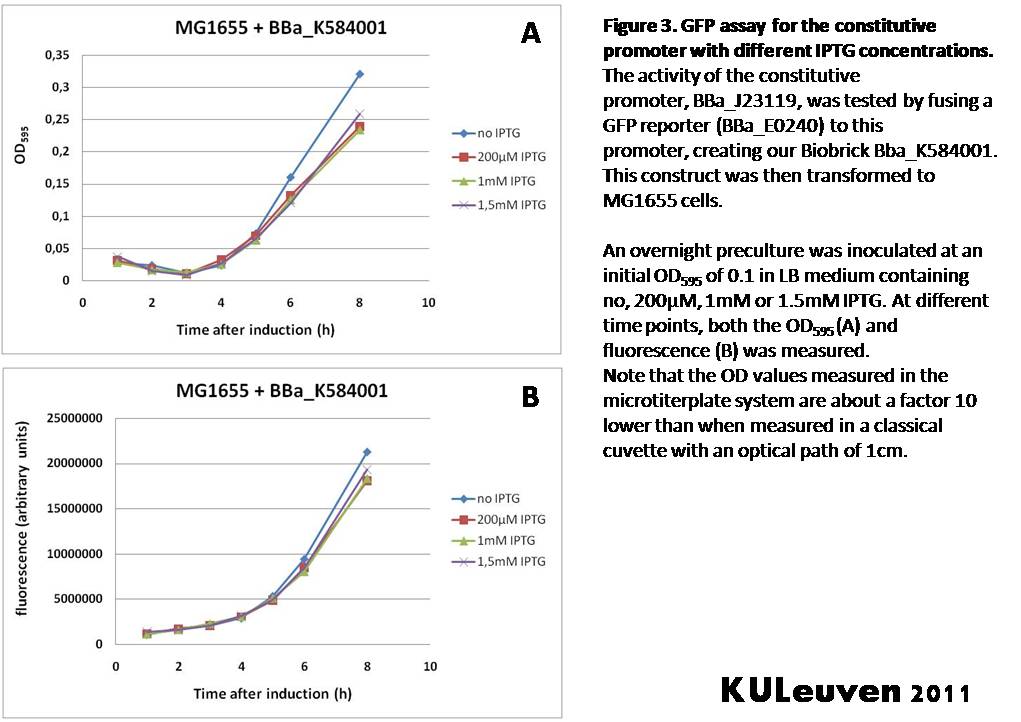
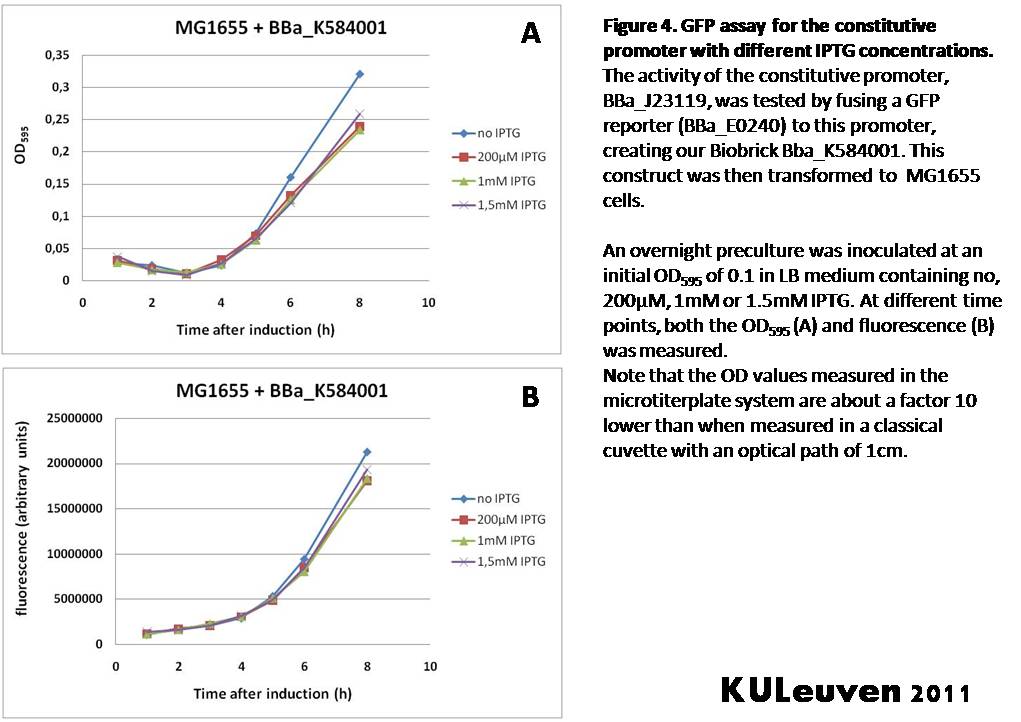
We tested the activity both in a TOP10F’ (figure 1 & 3) as well as a MG1655 (figure 2 & 4) E.coli strain background. For more information on E.coli strain descriptions, we recommend the following website: [http://openwetware.org/wiki/E._coli_genotypes E.Coli_Genotypes].</p> | We tested the activity both in a TOP10F’ (figure 1 & 3) as well as a MG1655 (figure 2 & 4) E.coli strain background. For more information on E.coli strain descriptions, we recommend the following website: [http://openwetware.org/wiki/E._coli_genotypes E.Coli_Genotypes].</p> | ||
Revision as of 07:03, 21 September 2011
Constitutive promoter + GFP generator
This part was designed to test the functionality of the constitutive promotor region.
Characterization
We fused the constitutive promoter to a GFP reporter, and assayed the promoter’s activity after addition of different amounts of arabinose and IPTG. As such these results can serve as a control for the results obtained for the arabinose-inducible BBa_K584000 and IPTG-inducible BBa_K584002 bricks. We tested the activity both in a TOP10F’ (figure 1 & 3) as well as a MG1655 (figure 2 & 4) E.coli strain background. For more information on E.coli strain descriptions, we recommend the following website: [http://openwetware.org/wiki/E._coli_genotypes E.Coli_Genotypes].
Apart from a small growth defect and somewhat lower fluorescence at the 8 hour time point after the addition of 2% arabinose, the activity of the constitutive promoter in TOP10F’ cells is unaffected by arabinose addition, as seen in Figure1. For the MG1655 cells (Figure 2), however, we see clear effects of adding arabinose at concentrations of 0.2% or 2%, with promoter activities significantly lower than the control condition. This points to interference of high inducer levels with the activity of the promoter and the importance of metabolism of the inducer, as arabinose can be metabolized by MG1655 cells, but not by TOP10F’ cells.
For both TOP10F’ and MG1655 cells, the activity of the constitutive promoter is unaffected by the addition of IPTG (Figures 3 and 4).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 706