Difference between revisions of "Part:BBa K404119"
| (6 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K404119 short</partinfo> | <partinfo>BBa_K404119 short</partinfo> | ||
| − | [[Image:Freiburg10_Vectorplasmid composite 1.png| | + | {| style="color:black" cellpadding="6" cellspacing="1" border="2" align="right" |
| + | ! colspan="2" style="background:#66bbff;"|[https://parts.igem.org/Part:BBa_K404119 [AAV2]-left-ITR_pCMV_betaglobin_mVenus_hGH_[AAV2]-right-ITR ] | ||
| + | |- | ||
| + | ! colspan="2"|[[Image:Freiburg10_Vectorplasmid composite 1.png|300px]] | ||
| + | |- | ||
| + | |'''BioBrick Nr.''' | ||
| + | |[https://parts.igem.org/Part:BBa_K404119 BBa_K404119] | ||
| + | |- | ||
| + | |'''RFC standard''' | ||
| + | |[https://parts.igem.org/Help:Assembly_standard_10 RFC 10]<br/> | ||
| + | [https://parts.igem.org/Help:Assembly_standard_25 RFC 25] | ||
| + | |- | ||
| + | |'''Requirement''' | ||
| + | |pSB1C3<br> | ||
| + | |- | ||
| + | |'''Source''' | ||
| + | |pAAV_MCS provided by Stratagene | ||
| + | |- | ||
| + | |'''Submitted by''' | ||
| + | |[http://2010.igem.org/Team:Freiburg_Bioware FreiGEM 2010] | ||
| + | |} | ||
| + | <br> | ||
| + | [[Image:Freiburg10_Vectorplasmid composite 1.png|left|thumb|400px]]<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
<html> | <html> | ||
| Line 121: | Line 143: | ||
</head> | </head> | ||
<body lang="EN-US"> | <body lang="EN-US"> | ||
| − | <div class="WordSection1"> | + | <div class="WordSection1"><br> |
| − | < | + | <p class="MsoNormal"> <big><big><big><big><big><big><big><big><big><big><big><big><big><span |
| − | <p class="MsoNormal"> < | + | |
| − | < | + | |
style="font-size: 10pt; line-height: 115%;">Producing | style="font-size: 10pt; line-height: 115%;">Producing | ||
recombinant virus particles for therapeutical applications is, besides | recombinant virus particles for therapeutical applications is, besides | ||
| Line 137: | Line 157: | ||
search. Subsequently, the essential components for AAV-2 particle | search. Subsequently, the essential components for AAV-2 particle | ||
production | production | ||
| − | were extracted and redesigned to match the iGEM standard.</span></p> | + | were extracted and redesigned to match the iGEM standard.</span></big></big></big></big></big></big></big></big></big></big></big></big></big></p> |
| − | <p class="MsoNormal"><span | + | <p class="MsoNormal"><small><small><small><small><small><small><small><span |
| − | style="font-size: 10pt; line-height: 115%;">The provided | + | style="font-size: 10pt; line-height: 115%;"><big><small>The |
| + | provided | ||
tripartite system is independent of a superinfection of | tripartite system is independent of a superinfection of | ||
Adeno- or herpes | Adeno- or herpes | ||
| Line 150: | Line 171: | ||
the preformed capsids after production of single-stranded DNA. | the preformed capsids after production of single-stranded DNA. | ||
Consequently, | Consequently, | ||
| − | this | + | this plasm</small></big></span></small></small></small></small></small></small></small><small><small><small><small><small><small><small><span |
| + | style="font-size: 10pt; line-height: 115%;"><big><small>id | ||
| + | is known as the vector plasmid (pGOI). Promoter, <i>beta-globin</i> | ||
intron and the hGH terminator signal are flanked by the ITRs (ITRs, | intron and the hGH terminator signal are flanked by the ITRs (ITRs, | ||
BBa_K404100 | BBa_K404100 | ||
| Line 156: | Line 179: | ||
containing the desired gene of interest is cotransfected with the | containing the desired gene of interest is cotransfected with the | ||
RepCap plasmid | RepCap plasmid | ||
| − | ( | + | (BBa_K40</small></big></span></small></small></small></small></small></small></small><small><small><small><small><small><small><small><span |
| − | obtain | + | style="font-size: 10pt; line-height: 115%;"><big><small>4001, |
| + | BBa_K404002 or BBa_K404003) and the pHelper plasmid. To obtain | ||
the fully assembled vector plasmid, several assembly steps have to be | the fully assembled vector plasmid, several assembly steps have to be | ||
| − | performed. </span></p> | + | performed.</small> </big> </span></small></small></small></small></small></small></small></p> |
| − | <p class="MsoNormal">After assembly of plasmids containing | + | <p class="MsoNormal"><small>After assembly of |
| − | all required elements, | + | plasmids containing all required elements, |
vector plasmid functionality was confirmed in cell culture. AAV-293 | vector plasmid functionality was confirmed in cell culture. AAV-293 | ||
cells | cells | ||
| Line 170: | Line 194: | ||
and the tumor cell line HT1080 was transduced with the recombinant | and the tumor cell line HT1080 was transduced with the recombinant | ||
viral | viral | ||
| − | vectors encapsidating the gene of interest mVenus (BBa_I757008).</p> | + | vectors encapsidating the gene of interest mVenus (BBa_I757008).</small></p> |
| − | <p class="MsoNormal">In the beginning, the iGEM team | + | <p class="MsoNormal"><small>In the beginning, the |
| − | Freiburg_Bioware 2010 used a | + | iGEM team Freiburg_Bioware 2010 used a |
commercial vector plasmid, pAAV-MCS (Stratagene), to determine whether | commercial vector plasmid, pAAV-MCS (Stratagene), to determine whether | ||
virus | virus | ||
| Line 185: | Line 209: | ||
(abbreviated pSB1C3_mVenus, BBa_K404119). Fluorescence expression data | (abbreviated pSB1C3_mVenus, BBa_K404119). Fluorescence expression data | ||
obtained | obtained | ||
| − | by flow cytometry analysis are shown in Figure | + | by flow cytometry analysis are shown in Figure 1. |
Comparing mVenus expression of the reference plasmid and the | Comparing mVenus expression of the reference plasmid and the | ||
pSB1C3_mVenus plasmid revealed | pSB1C3_mVenus plasmid revealed | ||
| − | that biological functionality of the reassembled plasmid was preserved. | + | that biological functionality of the reassembled plasmid was preserved.</small></p> |
| − | </p> | + | <p class="MsoNormal"></p> |
| + | <p class="MsoNormal"><small></small> </p> | ||
<p class="MsoNormal"><span | <p class="MsoNormal"><span | ||
style="color: rgb(0, 176, 80);"></span></p> | style="color: rgb(0, 176, 80);"></span></p> | ||
<table class="MsoTableGrid" | <table class="MsoTableGrid" | ||
| − | style="border: medium none ; border-collapse: collapse; width: | + | style="border: medium none ; border-collapse: collapse; width: 644px; height: 935px;" |
border="1" cellpadding="0" cellspacing="0"> | border="1" cellpadding="0" cellspacing="0"> | ||
<tbody> | <tbody> | ||
| Line 217: | Line 242: | ||
style="border-style: none solid solid; border-color: -moz-use-text-color windowtext windowtext; border-width: medium 1pt 1pt; padding: 0cm 5.4pt; width: 481.1pt;" | style="border-style: none solid solid; border-color: -moz-use-text-color windowtext windowtext; border-width: medium 1pt 1pt; padding: 0cm 5.4pt; width: 481.1pt;" | ||
valign="top" width="641"> | valign="top" width="641"> | ||
| − | <p class="MsoCaption"><a name="_Ref275784576">Figure</a> | + | <p class="MsoCaption"><small><a |
| − | 1: Flow cytometry analysis of fluorescent protein expression in | + | name="_Ref275784576">Figure</a> 1: Flow cytometry |
| − | transduced HT1080 cells. <span style="font-weight: normal;">For | + | analysis of fluorescent protein expression in transduced HT1080 cells. <span |
| − | viral particle production, AAV-293 cells were transfected with the | + | style="font-weight: normal;">For viral particle production, |
| − | reassembled vector plasmid (BBa_K404119) or the reference plasmid, | + | AAV-293 cells were transfected with the reassembled vector plasmid |
| − | respectively.</span> A<span style="font-weight: normal;">: | + | (BBa_K404119) or the reference plasmid, respectively.</span> A</small><span |
| − | Gating non transduced cells (control); subcellular debris and cellular | + | style="font-weight: normal;"><small>: Gating non |
| − | aggreates can be distinguished from single cells by size, estimated | + | transduced cells (control); subcellular debris and cellular aggreates |
| − | forward scatter (FS Lin) and granularity, estimated side scatter (SS | + | can be distinguished from single cells by size, estimated forward |
| − | Lin) B: Non-transduced cells plotted against cells expressing mVenus | + | scatter (FS Lin) and granularity, estimated side scatter (SS Lin) B: |
| + | Non-transduced cells plotted against cells expressing mVenus | ||
(Analytical gate was set such that 1% or fewer of negative control | (Analytical gate was set such that 1% or fewer of negative control | ||
cells fell within the positive region (R5) C: Gating transduced cells | cells fell within the positive region (R5) C: Gating transduced cells | ||
| − | (R2 </span><span | + | (R2 </small></span><small><span |
style="font-family: "Cambria Math","serif"; font-weight: normal;">≙ | style="font-family: "Cambria Math","serif"; font-weight: normal;">≙ | ||
</span><span style="font-weight: normal;">R14) | </span><span style="font-weight: normal;">R14) | ||
| Line 250: | Line 276: | ||
style="font-weight: normal;">: Transduced cells plotted | style="font-weight: normal;">: Transduced cells plotted | ||
against cells expressing mVenus. R10 comprises transduced cells | against cells expressing mVenus. R10 comprises transduced cells | ||
| − | detected by mVenus fluorescence.</span> J<span | + | detected by mVenus fluorescence.</span> J</small><span |
| − | style="font-weight: normal;">: Overlay of non-transduced | + | style="font-weight: normal;"><small>: Overlay of |
| − | (red) and transduced (green) cells plotted against mVenus expression. </span></p> | + | non-transduced (red) and transduced (green) cells plotted against |
| + | mVenus expression.</small> </span></p> | ||
</td> | </td> | ||
<td></td> | <td></td> | ||
| Line 261: | Line 288: | ||
<p class="MsoNormal"> </p> | <p class="MsoNormal"> </p> | ||
<table class="MsoTableGrid" | <table class="MsoTableGrid" | ||
| − | style="border: medium none ; border-collapse: collapse; width: | + | style="border: medium none ; border-collapse: collapse; width: 640px; height: 413px;" |
border="1" cellpadding="0" cellspacing="0"> | border="1" cellpadding="0" cellspacing="0"> | ||
<tbody> | <tbody> | ||
| Line 268: | Line 295: | ||
style="border: 1pt solid windowtext; padding: 0cm 5.4pt; width: 481.1pt;" | style="border: 1pt solid windowtext; padding: 0cm 5.4pt; width: 481.1pt;" | ||
valign="top" width="641"> | valign="top" width="641"> | ||
| − | <p class=" | + | <p class="MsoCaption"><img alt="" |
| − | + | src="%5BImage:Freiburg10%20Quantification%20of%20mVenus%20fluorescence%20by%20flow%20cytometry%20analysis.png%5D"> <br> | |
| − | + | <img style="width: 481px; height: 381px;" alt="" | |
| − | src="%5BImage:Freiburg10%20Quantification%20of%20mVenus%20fluorescence%20by%20flow%20cytometry%20analysis.png%5D"> < | + | id="Diagramm 90" |
| − | + | src="https://static.igem.org/mediawiki/2010/8/8e/Freiburg10_Quantification_of_mVenus_fluorescence_by_flow_cytometry_analysis.png"><br> | |
| − | + | <small><a name="_Ref275784852">Figure</a> | |
| − | + | 2: Quantification of mVenus fluorescence by flow cytometry analysis. </small><span | |
| − | + | style="font-weight: normal;"><small>The data source is | |
| − | src="https://static.igem.org/mediawiki/2010/8/8e/Freiburg10_Quantification_of_mVenus_fluorescence_by_flow_cytometry_analysis.png">< | + | identical to Figure 1. Compared to the reference vector plasmid, |
| − | < | + | mVenus expression in HT1080 cells is enhanced in case of the assembled |
| − | 2: Quantification of mVenus fluorescence by flow cytometry analysis. <span | + | plasmid (pSB1C3_mVenus)</small> </span></p> |
| − | style="font-weight: normal;">The data source is identical to | + | |
| − | Figure | + | |
| − | in HT1080 cells is enhanced in case of the assembled | + | |
| − | (pSB1C3_mVenus) </span></p> | + | |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 295: | Line 318: | ||
</body> | </body> | ||
</html> | </html> | ||
| + | |||
| + | |||
Latest revision as of 12:42, 31 October 2010
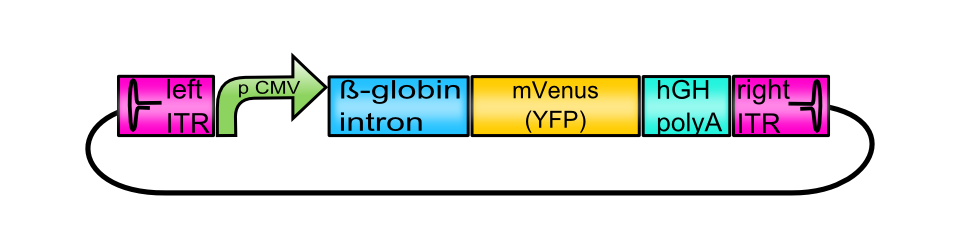
[AAV2]-left-ITR_pCMV_betaglobin_mVenus_hGH_[AAV2]-right-ITR
| [AAV2-left-ITR_pCMV_betaglobin_mVenus_hGH_[AAV2]-right-ITR ] | |
|---|---|
| |
| BioBrick Nr. | BBa_K404119 |
| RFC standard | RFC 10 |
| Requirement | pSB1C3 |
| Source | pAAV_MCS provided by Stratagene |
| Submitted by | [http://2010.igem.org/Team:Freiburg_Bioware FreiGEM 2010] |
Producing recombinant virus particles for therapeutical applications is, besides specific cell targeting, purification and quantification assays of AAV-2, one intention of the Virus Construction Kit provided by the iGEM team Freiburg_Bioware 2010. For obtaining a modular toolkit, the complex biological system of the Adeno-associated virus serotype 2 was examined by an exhaustive literature search. Subsequently, the essential components for AAV-2 particle production were extracted and redesigned to match the iGEM standard.
The provided tripartite system is independent of a superinfection of Adeno- or herpes simplex viruses since the genes encoding the required helper-proteins are co-transfected. Inside the eukaryotic host cell, the DNA sequence containing the inverted terminal repeats (ITRs) is extracted and later encapsidated into the preformed capsids after production of single-stranded DNA. Consequently, this plasmid is known as the vector plasmid (pGOI). Promoter, beta-globin intron and the hGH terminator signal are flanked by the ITRs (ITRs, BBa_K404100 and BBa_K404101) and regulate transgene expression. The vector plasmid containing the desired gene of interest is cotransfected with the RepCap plasmid (BBa_K404001, BBa_K404002 or BBa_K404003) and the pHelper plasmid. To obtain the fully assembled vector plasmid, several assembly steps have to be performed.
After assembly of plasmids containing all required elements, vector plasmid functionality was confirmed in cell culture. AAV-293 cells stably expressing the E1A and E1B proteins were transfected with three plasmids (pHelper, pRC, pGOI). Virus particles were harvested 72 hours post transfection and the tumor cell line HT1080 was transduced with the recombinant viral vectors encapsidating the gene of interest mVenus (BBa_I757008).
In the beginning, the iGEM team Freiburg_Bioware 2010 used a commercial vector plasmid, pAAV-MCS (Stratagene), to determine whether virus particle production by AAV-293 cells could be achieved. iGEM RFC 25 restriction enzyme sites were introduced and the fluorescent protein mVenus was subcloned into this plasmid. Subsequently, this plasmid was used as a reference and compared to the assembled vector plasmid pSB1C3_lITR_CMV_beta-globin_mVenus_hGH_rITR (abbreviated pSB1C3_mVenus, BBa_K404119). Fluorescence expression data obtained by flow cytometry analysis are shown in Figure 1. Comparing mVenus expression of the reference plasmid and the pSB1C3_mVenus plasmid revealed that biological functionality of the reassembled plasmid was preserved.
|
|
|
|
Figure 1: Flow cytometry analysis of fluorescent protein expression in transduced HT1080 cells. For viral particle production, AAV-293 cells were transfected with the reassembled vector plasmid (BBa_K404119) or the reference plasmid, respectively. A: Gating non transduced cells (control); subcellular debris and cellular aggreates can be distinguished from single cells by size, estimated forward scatter (FS Lin) and granularity, estimated side scatter (SS Lin) B: Non-transduced cells plotted against cells expressing mVenus (Analytical gate was set such that 1% or fewer of negative control cells fell within the positive region (R5) C: Gating transduced cells (R2 ≙ R14) (plasmids used for transfection: pGOI: pSB1C3_lITR_CMV_beta-globin_mVenus_hGH_rITR (pSB1C3_mVenus: BBa_K404119), pHelper, pRC. D: Transduced cells plotted against cells expressing mVenus. R10 comprises transduced cells detected by mVenus fluorescence. E: Overlay of non-transduced (red) and transduced (green). F: Gating non-transduced cells (control). G: Non-transduced cells plotted against cells expressing mVenus (R5). H: Gating transduced cells (R14 ≙ R2) (plasmids used for transfection: pGOI: pAAV_mVenus, pHelper). I: Transduced cells plotted against cells expressing mVenus. R10 comprises transduced cells detected by mVenus fluorescence. J: Overlay of non-transduced (red) and transduced (green) cells plotted against mVenus expression. |
|
|
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1319
Illegal AgeI site found at 2039 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 2441

 pSB1C3_mVenus
(BBa_K404119)
pSB1C3_mVenus
(BBa_K404119)

