Difference between revisions of "Part:BBa K404246"
| (2 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K404246 short</partinfo> | <partinfo>BBa_K404246 short</partinfo> | ||
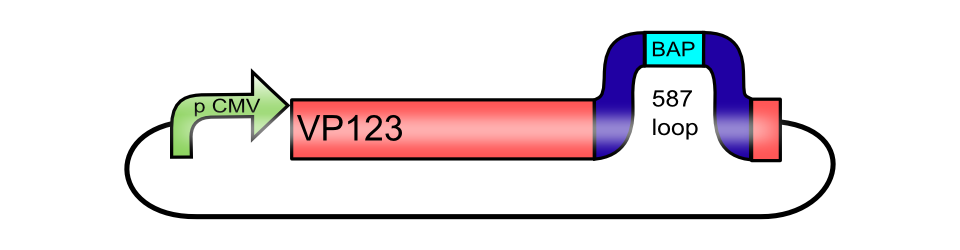
<br><b>The Biotinylation Acceptor peptide motif, inserted into the 587 loop of [[Part:BBa_K404007|pCMV_[AAV2]-VP123]]</b> | <br><b>The Biotinylation Acceptor peptide motif, inserted into the 587 loop of [[Part:BBa_K404007|pCMV_[AAV2]-VP123]]</b> | ||
| + | {| style="color:black" cellpadding="6" cellspacing="1" border="2" align="left" | ||
| + | ! colspan="2" style="background:#66bbff;"|[https://parts.igem.org/Part:BBa_K404246 [pCMV_[AAV2]-VP123ex(ViralBrick-587-BAP] | ||
| + | |- | ||
| + | |'''BioBrick Nr.''' | ||
| + | |[https://parts.igem.org/Part:BBa_K404246 BBa_K404246] | ||
| + | |- | ||
| + | |'''RFC standard''' | ||
| + | |[https://parts.igem.org/Help:Assembly_standard_10 RFC 10] | ||
| + | |- | ||
| + | |'''Requirement''' | ||
| + | |pSB1C3<br> | ||
| + | |- | ||
| + | |'''Source''' | ||
| + | |pAAV_MCS provided by Stratagene | ||
| + | |- | ||
| + | |'''Submitted by''' | ||
| + | |[http://2010.igem.org/Team:Freiburg_Bioware FreiGEM 2010] | ||
| + | |} | ||
| + | <br /><br /><br /><br /><br /><br /><br /><br /> | ||
[[Image:Freiburg10_pCMV_VP123 587-BAP.png|thumb|center|480px]]<br> | [[Image:Freiburg10_pCMV_VP123 587-BAP.png|thumb|center|480px]]<br> | ||
| + | |||
| + | The AAV capsid consists of 60 capsid protein subunits. The three cap proteins VP1, VP2, and VP3 are encoded in an overlapping reading frame. Arranged in a stoichiometric ratio of 1:1:10, they form an icosahedral symmetry. The mRNA encoding for the cap proteins is transcribed from p40 and alternative spliced to minor and major products. Alternative splicing and translation initiation of VP2 at a nonconventional ACG initiation codon promote the expression of VP1, VP2 and VP3. The VP proteins share a common C terminus and stop codon, but begin with a different start codon. The N termini of VP1 and VP2 play important roles in infection and contain motifs that are highly homologous to the phospholipase A2 (PLA2) domain and nuclear localization signals (BR)(+). | ||
| + | <br /> | ||
| + | <br> | ||
| + | CMV promoter is derived from human Cytomegalovirus, which belongs to Herpesvirus group. All family members share the ability to remain in latent stage in the human body. CMV is located upstream of immediate-early gene. However, CMV promoter is an example of widely used promoters and is present in mammalian expression vectors. The advantage of CMV is the high-level constitutive expression in mostly all human tissues [Fitzsimons et al., 2002]. | ||
| + | <br> | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
Latest revision as of 16:11, 13 January 2011
pCMV_[AAV2]-VP123ex (ViralBrick-587-BAP)
The Biotinylation Acceptor peptide motif, inserted into the 587 loop of pCMV_[AAV2]-VP123
| [pCMV_[AAV2-VP123ex(ViralBrick-587-BAP] | |
|---|---|
| BioBrick Nr. | BBa_K404246 |
| RFC standard | RFC 10 |
| Requirement | pSB1C3 |
| Source | pAAV_MCS provided by Stratagene |
| Submitted by | [http://2010.igem.org/Team:Freiburg_Bioware FreiGEM 2010] |
The AAV capsid consists of 60 capsid protein subunits. The three cap proteins VP1, VP2, and VP3 are encoded in an overlapping reading frame. Arranged in a stoichiometric ratio of 1:1:10, they form an icosahedral symmetry. The mRNA encoding for the cap proteins is transcribed from p40 and alternative spliced to minor and major products. Alternative splicing and translation initiation of VP2 at a nonconventional ACG initiation codon promote the expression of VP1, VP2 and VP3. The VP proteins share a common C terminus and stop codon, but begin with a different start codon. The N termini of VP1 and VP2 play important roles in infection and contain motifs that are highly homologous to the phospholipase A2 (PLA2) domain and nuclear localization signals (BR)(+).
CMV promoter is derived from human Cytomegalovirus, which belongs to Herpesvirus group. All family members share the ability to remain in latent stage in the human body. CMV is located upstream of immediate-early gene. However, CMV promoter is an example of widely used promoters and is present in mammalian expression vectors. The advantage of CMV is the high-level constitutive expression in mostly all human tissues [Fitzsimons et al., 2002].
Usage and Biology

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 2396
Illegal XhoI site found at 698
Illegal XhoI site found at 884 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 665
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 2982
Illegal SapI site found at 1833