Difference between revisions of "Part:BBa K4150001"
Jacky CHIN (Talk | contribs) |
Jacky CHIN (Talk | contribs) |
||
| (17 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K4150001 short</partinfo> | <partinfo>BBa_K4150001 short</partinfo> | ||
| − | The part is composed of a T7 promoter ([[Part:BBa_I719005]]) and a strong RBS containing the T7 g10 leader sequence ([[Part:BBa_K4150000]]) | + | <br> |
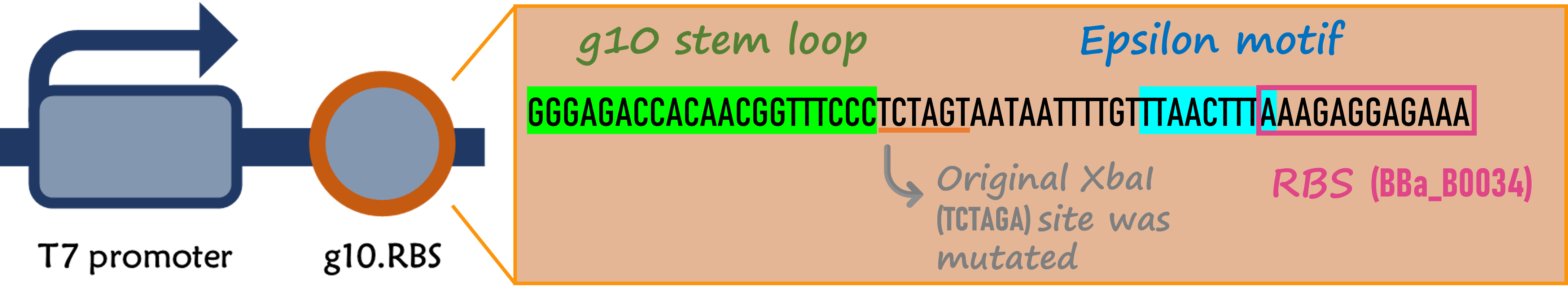
| + | <span style="color:#00000000">This</span>The part is composed of a T7 promoter ([[Part:BBa_I719005]]) and a strong RBS containing the T7 g10 leader sequence ([[Part:BBa_K4150000]]) | ||
| + | |||
| + | <br> | ||
| + | |||
| + | [[File:T--Mingdao--2022 Registry 01.png|700px|center]] | ||
| + | |||
| + | <br> | ||
| + | |||
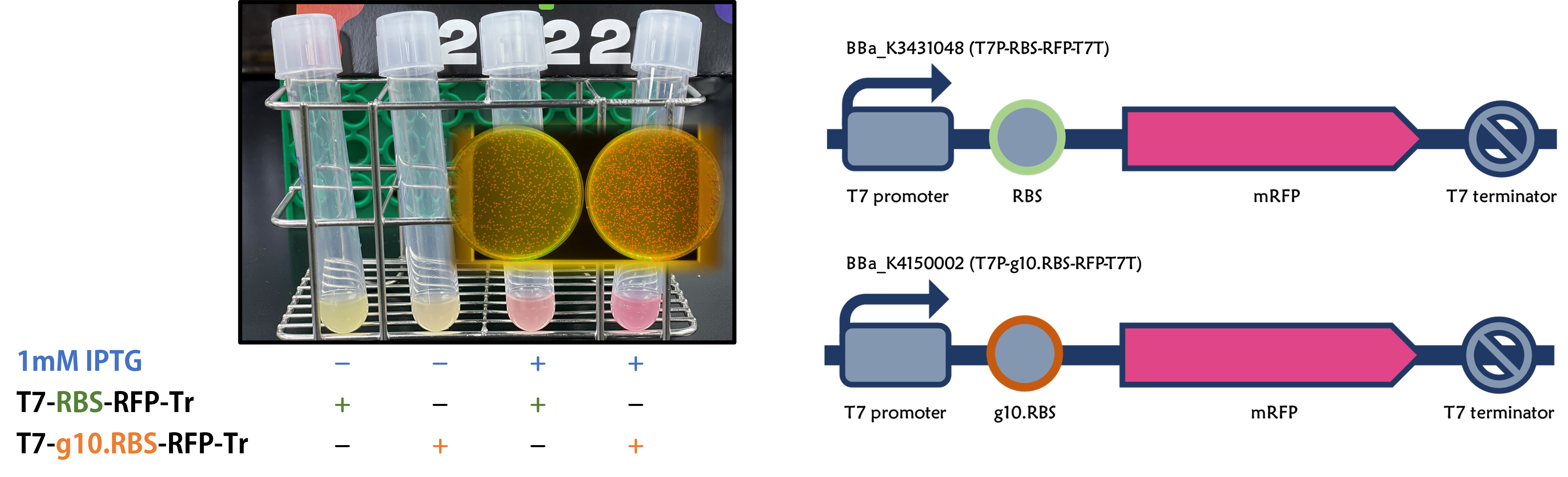
| + | <span style="color:#00000000">This</span>We replaced the RBS with g10.RBS in BioBrick [[Part:BBa_K3431048]] (T7-RBS-RFP-Tr) to create an improved BioBrick [[Part:BBa_K4150002]] (T7-g10.RBS-RFP-Tr). These two plasmids were transformed into E. coli BL21. In the presence of IPTG, colonies on the LB agar plate and cultures in the LB broth both showed darker red colors for RFP gene expression driven by T7 promoter with g10 leader sequence compared to the original RBS without g10 leader sequence (Fig.1). However, the E. coli BL21 with T7-g10.RBS-RFP-Tr had a higher background level in the absence of IPTG. | ||
| + | |||
| + | <br> | ||
| + | |||
| + | |||
| + | [[File:T--Mingdao--2022 Registry 02.png|950px|center]] | ||
| + | |||
| + | Figure 1. The plasmids as indicated were transformed into E. coli BL21. The E. coli were grown on LB agar plate supplemented with 20 μg/mL of chloramphenicol and 1mM of IPTG. One of the colonies was cultured in LB broth with 34 μg/mL of chloramphenicol in the absence or in the presence of 1mM of IPTG. Inset panel: The plates were observed under a blue LED light box. | ||
| + | |||
| + | |||
| + | <br><br> | ||
| + | |||
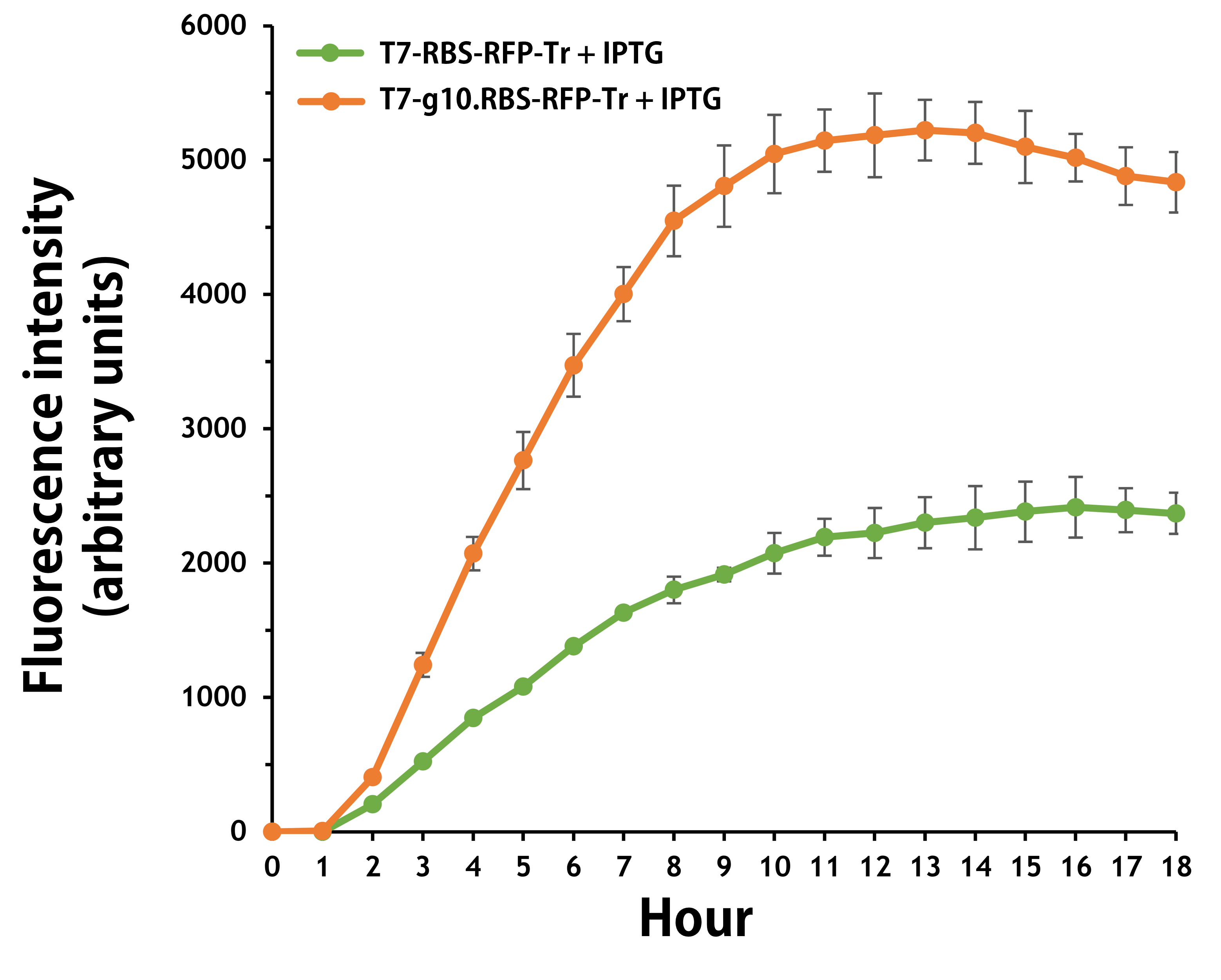
| + | <span style="color:#00000000">This</span>Furthermore, the transformed E. coli with T7-g10.RBS-RFP-Tr compared to T7-RBS-RFP-Tr expressed RFP at faster and increased (up to 2.5-fold) fluorescence intensity levels in a time-dependent manner (Fig. 2). The above data demonstrated the g10 leader sequence in front of RBS can significantly enhance gene expression. | ||
| + | |||
| + | <br> | ||
| + | [[File:T--Mingdao--2022 Engineering Success photo 10.png|500px|left]] | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | Figure 2. The plasmids as indicated were transformed into E. coli BL21. The E. coli were grown in LB broth supplemented with 34 μg/mL of chloramphenicol and 1mM of IPTG for 18 hours. The RFP expression levels were read at ex/em = 584/607 nm in a kinetic mode by Synergy H1 Hybrid Multi-Mode Reader - BioTek Instruments (Agilent Technologies, Inc.). The values of fluorescence intensity were presented by the data with IPTG induction minus the data without induction as background levels. | ||
| + | |||
| + | <br><br> | ||
| + | |||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 05:59, 19 August 2022
T7P-g10.RBS
ThisThe part is composed of a T7 promoter (Part:BBa_I719005) and a strong RBS containing the T7 g10 leader sequence (Part:BBa_K4150000)
ThisWe replaced the RBS with g10.RBS in BioBrick Part:BBa_K3431048 (T7-RBS-RFP-Tr) to create an improved BioBrick Part:BBa_K4150002 (T7-g10.RBS-RFP-Tr). These two plasmids were transformed into E. coli BL21. In the presence of IPTG, colonies on the LB agar plate and cultures in the LB broth both showed darker red colors for RFP gene expression driven by T7 promoter with g10 leader sequence compared to the original RBS without g10 leader sequence (Fig.1). However, the E. coli BL21 with T7-g10.RBS-RFP-Tr had a higher background level in the absence of IPTG.
Figure 1. The plasmids as indicated were transformed into E. coli BL21. The E. coli were grown on LB agar plate supplemented with 20 μg/mL of chloramphenicol and 1mM of IPTG. One of the colonies was cultured in LB broth with 34 μg/mL of chloramphenicol in the absence or in the presence of 1mM of IPTG. Inset panel: The plates were observed under a blue LED light box.
ThisFurthermore, the transformed E. coli with T7-g10.RBS-RFP-Tr compared to T7-RBS-RFP-Tr expressed RFP at faster and increased (up to 2.5-fold) fluorescence intensity levels in a time-dependent manner (Fig. 2). The above data demonstrated the g10 leader sequence in front of RBS can significantly enhance gene expression.
Figure 2. The plasmids as indicated were transformed into E. coli BL21. The E. coli were grown in LB broth supplemented with 34 μg/mL of chloramphenicol and 1mM of IPTG for 18 hours. The RFP expression levels were read at ex/em = 584/607 nm in a kinetic mode by Synergy H1 Hybrid Multi-Mode Reader - BioTek Instruments (Agilent Technologies, Inc.). The values of fluorescence intensity were presented by the data with IPTG induction minus the data without induction as background levels.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 34