Difference between revisions of "Part:BBa K3610003"
(→Plant Immunity Based Biosensing) |
|||
| (30 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
This part is a version of the BAK1 receptor from <i>A. thaliana</i> ([[Part:BBa_K3610001]] for full length) without the intracellular kinase domain and without the native signal sequence. | This part is a version of the BAK1 receptor from <i>A. thaliana</i> ([[Part:BBa_K3610001]] for full length) without the intracellular kinase domain and without the native signal sequence. | ||
| + | <html> | ||
| + | </p> | ||
| + | </html> | ||
| + | __TOC__ | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | [[File:T--UZurich--BAK1 ecto.png|300px|thumb|left|Figure 1: BAK1 ectodomain protein structure (Uniprot Q94F62)]] | ||
| − | The BRI1-associated receptor kinase (BAK1) is a | + | |
| + | The BRI1-associated receptor kinase (BAK1) is a leucine-rich repeat receptor kinase (LRR-RK) which interacts with multiple other LRR-RKs with different functions in hormone signalling and defense response. BAK1 localizes at the plasma membrane and the endosome. The BAK1 protein forms a structure with an extracellular domain with leucine-rich repeats, a single pass transmembrane domain and an intracellular domain with a kinase function. | ||
Among others, BAK1 interacts with the LRR-RKs EF-Tu receptor (EFR), Flagellin sensing 2 (FLS2) and cold-shock protein receptor (CORE), all of which are pathogen recognition receptors (PRR) in brassicaceae plants. Upon binding of a microbe-associated molecular pattern at the LRR domain of the PRR, BAK1 forms a heterodimer with the PRR which triggers a phosphorylation cascade, leading to upregulation of defense mechanisms. | Among others, BAK1 interacts with the LRR-RKs EF-Tu receptor (EFR), Flagellin sensing 2 (FLS2) and cold-shock protein receptor (CORE), all of which are pathogen recognition receptors (PRR) in brassicaceae plants. Upon binding of a microbe-associated molecular pattern at the LRR domain of the PRR, BAK1 forms a heterodimer with the PRR which triggers a phosphorylation cascade, leading to upregulation of defense mechanisms. | ||
| − | This part includes only the ectodomain and the transmembrane domain. The intracellular kinase domain was cleaved off. Additionally, the natural signal peptide sequence, which is necessary for localization at the cell membrane in the plant, has been cleaved off as well. Therefore, this part needs to be used together with a signal peptide from the organism in which the protein is expressed. | + | This part includes only the ectodomain and the transmembrane domain. The intracellular kinase domain was cleaved off. Additionally, the natural signal peptide sequence, which is necessary for localization at the cell membrane in the plant, has been cleaved off as well. Therefore, this part needs to be used together with a signal peptide from the organism in which the protein is expressed.<br /> |
| + | <br><br><br><br> | ||
| − | ==Plant Immunity Based Biosensing== | + | ==Plant Immunity Based Biosensing UZürich 2020== |
For visualizing the interaction of BAK1 with other plant receptors the cytoplasmic domain can be replaced with a split fluorescent protein or another protein that generates a visual output. For our iGEM project, we designed a system to use this mechanism for detection of bacterial epitopes in water. | For visualizing the interaction of BAK1 with other plant receptors the cytoplasmic domain can be replaced with a split fluorescent protein or another protein that generates a visual output. For our iGEM project, we designed a system to use this mechanism for detection of bacterial epitopes in water. | ||
We used this part for multiple experiments. | We used this part for multiple experiments. | ||
| Line 23: | Line 30: | ||
We expressed this protein in <i>S. cerevisiae</i> to test two things. First of all, we wanted to see whether the protein gets expressed at all, and secondly, we were interested in seeing, whether the secretion signal from yeast and the receptor without the intracellular kinase domain would be sufficient for localization at the cell periphery. | We expressed this protein in <i>S. cerevisiae</i> to test two things. First of all, we wanted to see whether the protein gets expressed at all, and secondly, we were interested in seeing, whether the secretion signal from yeast and the receptor without the intracellular kinase domain would be sufficient for localization at the cell periphery. | ||
| − | In a first step we inserted the single fragments making up this part into a plasmid with a gentamycin-3-acetyltransferase gene and transformed <i>E. coli</i> (DH10alpha) with the plasmids for amplification. In the next step we assembled the fragments in a plasmid with a spectinomycin acetyltransferase and amplified the plasmids again in the same <i>E. coli</i> strain. For this step we applied the techniques of Golden Gate Cloning to get the fragments in the right order into the plasmid. The restriction enzyme we chose was BsaI. For expressing this part consisting of YFP and the receptor protein, we initially intended to use promoters of different | + | In a first step we inserted the single fragments making up this part into a plasmid with a gentamycin-3-acetyltransferase gene and transformed <i>E. coli</i> (DH10alpha) with the plasmids for amplification. In the next step we assembled the fragments in a plasmid with a spectinomycin acetyltransferase and amplified the plasmids again in the same <i>E. coli</i> strain. For this step we applied the techniques of Golden Gate Cloning to get the fragments in the right order into the plasmid. The restriction enzyme we chose was BsaI. For expressing this part consisting of YFP and the receptor protein, we initially intended to use promoters of different strengths to get more quantitative data. Finally, we got the construct in a plasmid with a truncated version of the ADH1 promoter from <i>S. cerevisiae</i>. For termination, this part has the terminator sequence of the enolase 2 protein from <i>S. cerevisiae</i>. The plasmid also contained the TRP1 gene, which encodes phosphoribosylanthranilate isomerase, an enzyme that catalyzes the third step in tryptophan biosynthesis. This enabled us to use the same plasmid for expression in <i>S. cerevisiae</i>. We prepared a medium containing YNB and free amino acids, without tryptophan. <i>S. cerevisiae</i> cells (AP4) were transfected with the plasmid and then plated on the selective medium. |
| + | |||
| + | ====Microscopy==== | ||
After successful transformation of yeast cells we checked for expression of the protein under a confocal microscope. | After successful transformation of yeast cells we checked for expression of the protein under a confocal microscope. | ||
| − | [[File:T--UZurich-- | + | [[File:T--UZurich--Control.png|800px|thumb|none|Figure 2: Control]] |
| − | [[File:T--UZurich-- | + | [[File:T--UZurich--eBAK.png|800px|thumb|none|Figure 3: cells transformed with eBAK fused to YFP, increased fluorescence]] |
| − | Confocal microscopy confirmed increased fluorescence in the <i>S. cerevisiae</i> cells that had been previously transfected with plasmids containing BAK1 ectodomain fused to YFP. This increased fluorescence indicates expression of our genes. | + | Confocal microscopy confirmed increased fluorescence in the <i>S. cerevisiae</i> cells that had been previously transfected with plasmids containing BAK1 ectodomain fused to YFP. This increased fluorescence indicates expression of our genes. As we were particularily interested in seeing where within the cell the receptor would get localized, we stained the membrane with the popular stain FM4-64 and then examined the samples again with confocal microscopy. |
| − | + | ||
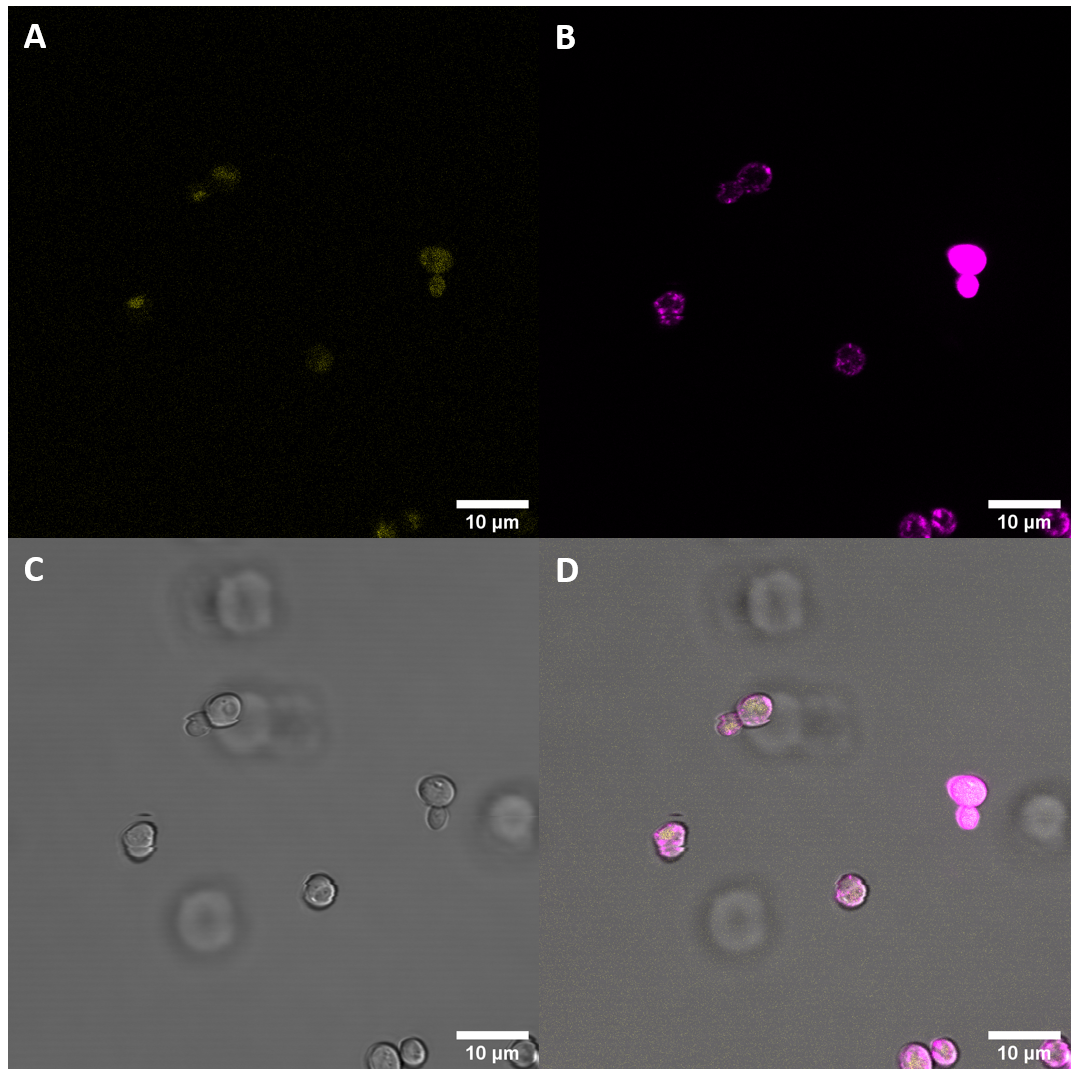
| + | [[File:T--UZurich--UT Membrane Stain.png|440px|thumb|left|Figure 4: Untransformed Control,(A) : YFP, (B) : FM4-64, (C): light field. (D): merge.Imaging of untransfected S. cerevisiae cells reveals hardly any fluorescence within the YFP spectrum]]<br> | ||
| + | [[File:T--UZurich--eBAK Membrane Stain.png|440px|thumb|none|Figure 5: eBAK <i>S. cerevisiae</i>: (A) : YFP, (B) : FM4-64, (C) : light field. (D): merge.Most eBAK cells show strong yellow fluorescence with only diffuse structures. | ||
| + | About 1% of the cells, clear co-localization with FM4-64 in a ring structure indicate integration into the PM.]] | ||
| + | |||
| + | This additional imaging experiment revealed that the fluorescent protein is in part localized at the cell periphery. This is in alignment with our expectations as our construct includes a secretion signal protein and a receptor coding protein with the transmembrane domain. | ||
These results suggest that the secretion peptide fused to the receptor ectodomain, including the transmembrane domain can be expressed in <i>S. cerevisiae</i> and that the components are sufficient for localization at the cell membrane. | These results suggest that the secretion peptide fused to the receptor ectodomain, including the transmembrane domain can be expressed in <i>S. cerevisiae</i> and that the components are sufficient for localization at the cell membrane. | ||
| + | |||
| + | This observation is already exciting and underlines plant PRR's potential as tools for Synthetic Biology. | ||
| + | |||
| + | ====Spectrometry==== | ||
| + | |||
| + | In addition to analyzing the cells with a microscope, we conducted a fluorescence assay with a plate reader. We conducted this experiment for multiple receptors at the same time. This way we were able to compare the expression levels of differnt versions of the BAK1 receptor. | ||
| + | For each receptor we tried to isolate three different biological samples, however, not all cells grew. | ||
| + | Ultimately, we only had two samples for the following <i>S. cerevisiae</i> cells: untransformed (Control), transformed with BAK1 ectodomain fused to YFP (eBAK) and the CORE ectodomain fused to YFP (eCORE). | ||
| + | For the BAK1 with and without the native signal peptide fused to YFP (BAK+ and BAK-) and the EFR ectodomain fused to YFP (eEFR), we had samples from three different colonies. | ||
| + | For each biological replicate, the optical density at absorbance of 600 nm (OD600) and the fluorescence levels were measured three times. | ||
| + | |||
| + | {| class="wikitable" width="95%" | ||
| + | |- | ||
| + | ! scope="col", colspan ="10"| measured OD600 values (OD) | ||
| + | |- | ||
| + | ! scope="col"| | ||
| + | ! scope="col", colspan="3"| Replicate 1 | ||
| + | ! scope="col", colspan="3"| Replicate 2 | ||
| + | ! scope="col", colspan="3"| Replicate 3 | ||
| + | |- | ||
| + | ! scope="row"| Blank | ||
| + | |0,08200000226||0,08200000226||0,08389999717|||||||||||| | ||
| + | |- | ||
| + | !Control | ||
| + | |0,3806000054||0,3747999966||0,4221999943||0,1316999942||0,131400004||0,1176000014|||||| | ||
| + | |- | ||
| + | !BAK+ | ||
| + | |0,4943000078||0,4638999999||0,4514000118||0,5781000257||0,5253999829||0,5799999833||0,2615999877||0,2171999961||0,2011999935 | ||
| + | |- | ||
| + | !BAK- | ||
| + | |1,417099953||1,365499973||1,368899941||0,6305999756||0,5633999705||0,6216999888||0,896600008||0,7882999778||0,8032000065 | ||
| + | |- | ||
| + | !eBAK | ||
| + | |1,009699941||0,8404999971||0,8934999704||0,2653000057||0,2368000001||0,2592999935|||||| | ||
| + | |- | ||
| + | !eCORE | ||
| + | |1,021499991||0,8616999984||0,9178000093||0,826300025||0,6888999939||0,7401999831|||||| | ||
| + | |- | ||
| + | !eEFR | ||
| + | |1,379699945||1,322700024||1,333500028||1,035899997||1,014000058||0,9526000023||0,4860999882||0,3797000051||0,3829999864 | ||
| + | |} | ||
| + | |||
| + | The following settings were applied for fluorescence measurements: | ||
| + | |||
| + | {| | ||
| + | |Mode: ||Fluorescence Top Reading | ||
| + | |- | ||
| + | |Excitation Wavelength: ||485 nm | ||
| + | |- | ||
| + | |Emission Wavelength: ||535 nm | ||
| + | |- | ||
| + | |Excitation Bandwidth: ||20 nm | ||
| + | |- | ||
| + | |Emission Bandwidth: ||25 nm | ||
| + | |- | ||
| + | |Temperature: ||22.3°C | ||
| + | |} | ||
| + | |||
| + | {| class="wikitable" width="95%" | ||
| + | |- | ||
| + | ! scope="col", colspan ="10"| Fluorescence Top Reading (FTR) | ||
| + | |- | ||
| + | ! scope="col"| | ||
| + | ! scope="col", colspan="3"| Replicate 1 | ||
| + | ! scope="col", colspan="3"| Replicate 2 | ||
| + | ! scope="col", colspan="3"| Replicate 3 | ||
| + | |- | ||
| + | ! scope="row"| Blank | ||
| + | |1297||1282||1322|||||||||||| | ||
| + | |- | ||
| + | !Control | ||
| + | |2684||2474||2634||1852||1792||1750|||||| | ||
| + | |- | ||
| + | !BAK+ | ||
| + | |3038||2813||2760||2836||2493||2788||2084||2072||2067 | ||
| + | |- | ||
| + | !BAK- | ||
| + | |35794||30319||31424||10792||9097||10517||22609||20227||21220 | ||
| + | |- | ||
| + | !eBAK | ||
| + | |26455||19828||21613||6614||5507||6229|||||| | ||
| + | |- | ||
| + | !eCORE | ||
| + | |10709||8382||9339||8957||7062||7735|||||| | ||
| + | |- | ||
| + | !eEFR | ||
| + | |43125||37782||39589||25641||24668||22517||12410||9054||9027 | ||
| + | |} | ||
| + | |||
| + | After measurement of the optical density and the fluorescence, the data were <i>blank corrected</i> (the average of the three blank measurements was subtracted from each measurement value). | ||
| + | |||
| + | The average of each of the three (or two) samples was calculated. From these values, the average was taken again. | ||
| + | |||
| + | After this step, we normalized the fluorescent output for OD600 (FTR/OD). | ||
| + | The results of these calculations are displayed in the table below. | ||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | !Control | ||
| + | !BAK+ | ||
| + | !BAK- | ||
| + | !eBAK | ||
| + | !eCORE | ||
| + | !eEFR | ||
| + | |- | ||
| + | |4185,221063||9731,614266||26067,19254||28118,24739||3712,946478||23379,84399 | ||
| + | |} | ||
| + | |||
| + | If we set the values for the Control to 1 (Control = 1), then we get the fluorescence levels relative to the control, which is again diplayed in the table below. | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | !Control | ||
| + | !eCORE | ||
| + | !eEFR | ||
| + | !BAK- | ||
| + | !BAK+ | ||
| + | !eBAK | ||
| + | |- | ||
| + | |1||0,8871565975||5,586286516||6,228390841||2,325233033||6,718461693 | ||
| + | |} | ||
| + | |||
| + | <br> | ||
| + | [[File:T--UZurich--Spectrometer1.png|500px|none|left|Figure 6: Fluorescence values standardized for OD600 of the different receptors (C=Control). Cells with BAK+ showed only weak fluorescence, while BAK-, eBAK and eEFR showed a strong increase in the fluorescence levels. CORE did not display any increase when compared with untreated <i>S. cerevisiae</i> cells (autofluorescence).]] | ||
| + | <br /> | ||
| + | Results from the plate reader and from confocal microscopy were in alignment. Measurement with the fluorometer suggested increased expression of YFP when <i>S. cerevisiae</i> cells are transfected with a plasmid that contains the BAK1 ectodomain fused to YFP. These results together strongly imply, that the ectodomain of the plant PRR BAK1 from <i>A. thaliana</i>, which has been fused to the secretion signal from the alpha-Factor, is expressed in yeast and also gets localized at the cell membrane. | ||
| + | |||
| + | ====Flow Cytometry==== | ||
| + | It has been important to us to examine a sample with different approaches simultaneously, which is why we were eager to also measure fluorescence intensity by flow cytometry. In a first phase, 100,000 cells were measured from each biological replicate (488/530 FITC channel in a BD FACSCanto II flow cytometer). | ||
| + | In the next phase, the biological replicates for each construct were pooled together and 200,000 cells from each sample were measured. | ||
| + | |||
| + | [[File:T--UZurich--FACs.png|600px|thumb|none|Figure 7: Left: single biological replicates. right: pooled samples. With the exception of the construct with eCORE, cells transfected with our constructs showed considerably higher overall fluorescence intensities than the negative control.]] | ||
| + | |||
| + | Flow catometry provided further evidence for expression of the construct in <i>S. cerevisiae</i>. Cells transfected with plasmids containing BAK- showed significantly increased fluorescence intensities when examined as single biological replicates, as well as when the replicates were pooled together in one sample. | ||
===Expression with split-mCherry=== | ===Expression with split-mCherry=== | ||
| − | For another part of our project, we fused the ectodomain to the N-terminal domain of the split-mCherry protein instead of YFP. The goal was to coexpress this construct ([[Part: | + | For another part of our project, we fused the ectodomain to the N-terminal domain of the split-mCherry protein instead of YFP. The goal was to coexpress this construct ([[Part:BBa_K3610034]]) with one of the plant PRRs that show ligand-dependent interaction with BAK1, like EFR. We managed to design a construct consisting of the ectodomain of EFR which was fused to a secretion signal and the C-terminal domain of mCherry ([[Part:BBa_K3610039]]. |
| − | Coexpressed in <i>S. cerevisiae</i>, these two parts are able to interact upon ligand-binding. | + | Coexpressed in <i>S. cerevisiae</i>, these two parts are able to interact upon ligand-binding. The goal of our experiments was to test, whether ligand-dependent dimerization of the mCherry protein would be sufficient to drive dimerization of the split-mCherry parts. |
| + | |||
| + | During our project, we achieved coexpression of [[Part:BBa_K3610034]] with [[Part:BBa_K3610042]], which encodes the ectodomain of the plant PRR target receptor EFR fused to the C-terminal domain of mCherry in <i>S. cerevisiae</i>. | ||
| + | |||
| + | ====Fluorescence Assay==== | ||
| + | To test, whether the EFR ligand elf18 would drive dimerization of the receptors, we conducted an experiment, during which we compared the fluorescent response of the yeast. | ||
| + | We therefore conducted an assay with a fluorometer, a plate reader of the type <i>Synergy H1</i> (λEX = 587nm and λEM = 610nm). We eamined untransformed cells as a control (UT) and cells which had been previously transfected with plasmids containting the parts [[Part:BBa_K3610034]] and [[Part:BBa_K3610042]] (eEFR). All samples had been adjusted to OD600 = o0.5 before the plate reader experiment.<br> | ||
| + | The following samples were prepared: | ||
| + | *UT | ||
| + | *UT, elf18 added | ||
| + | *eEFR | ||
| + | *eEFR, elf18 added | ||
| + | |||
| + | [[File:T--UZurich--Kinetics mCherry.png|600px|thumb|none|Figure 8: Comparison of average luminescence measurements over time for different samples.]] | ||
| + | |||
| + | Surprisingly, fluorescence levels did increase when the bacterial elicitor was present, however, this was the case for both types of samples. We must be, however, aware of the fact that the variance was rather large and this one time assay does not provide enough data to confirm a significant effect of the bacterial elicitor. | ||
| + | What was an even bigger surprise is that the measured fluorescence intensities were higher for the samples containing the untransformed yeast cells, both when the bacterial elicitor was present and when it had not been added. | ||
| + | The reasons for these results are unclear. It could be due to an error when adjusting the samples to the same OD600. A difference in the OD could lead to increased fluorescence levels. This would still not explain why the bacterial elicitor seemed to increase autofluorescence for the untransformed yeast cells. | ||
| + | |||
| + | It is clear that this deserves further examination. We propose to repeat the measurements to increase the data coverage and rule out errors during pipetting or labelling the samples. It should further be attempted to lower autofluorescence for yeast cells that resembles the fluorescence of mCherry. It further needs to examined, whether the same type of results are obtained when a different split-reporter is fused to the receptor domain (something which we were able to carry out during our project). | ||
===Expression with split-NanoLuc=== | ===Expression with split-NanoLuc=== | ||
For another part of our project, we fused the ectodomain to the LargeBit part of the split-NanoLuc protein instead of the intracellular kinase domain. The goal was to coexpress this construct ([[Part:BBa_K3610032]]) with one of the plant PRRs that show ligand-dependent interaction with BAK1, for example EFR. We managed to design a construct consisting of the ectodomain of EFR which was fused to a secretion signal and the SmallBit of NanoLuc ([[Part:BBa_K3610043]]. | For another part of our project, we fused the ectodomain to the LargeBit part of the split-NanoLuc protein instead of the intracellular kinase domain. The goal was to coexpress this construct ([[Part:BBa_K3610032]]) with one of the plant PRRs that show ligand-dependent interaction with BAK1, for example EFR. We managed to design a construct consisting of the ectodomain of EFR which was fused to a secretion signal and the SmallBit of NanoLuc ([[Part:BBa_K3610043]]. | ||
Coexpressed in <i>S. cerevisiae</i>, these two parts are able to interact upon ligand-binding. This dimerization will reconstitute the functionality of the NanoLuc protein, leading to a chemiluminescent output in the presence of furimazine. | Coexpressed in <i>S. cerevisiae</i>, these two parts are able to interact upon ligand-binding. This dimerization will reconstitute the functionality of the NanoLuc protein, leading to a chemiluminescent output in the presence of furimazine. | ||
| + | |||
| + | ====Luminescence Assay 1==== | ||
| + | We managed to coexpress the BAK1 ectodomain with LargeBit together with two different receptor ectodomains fused to the SmallBit part of NanoLuc, EFR (eEFR) and CORE (eCORE). | ||
| + | |||
| + | After transfection of <i>S. cerevisiae</i> cells, samples were prepared and examined with a luminometer. We used a plate reader of the type <i>Synergy H1</i>. For a more detailed view on data visite the page of the composite part: [[Part:BBa K3610038]]. | ||
| + | |||
| + | Samples were prepared: | ||
| + | *untransfected control (UT) | ||
| + | *eEFR and eBAK1 (eEFR) | ||
| + | *eCORE and eBAk1 (CORE) | ||
| + | |||
| + | each sample type was used for three types of measurements: | ||
| + | *no bacterial elicitor added | ||
| + | *csp22 added | ||
| + | *elf18 added | ||
| + | |||
| + | Applying all setups to each sample, we measured luminescence intensities for 9 different groups. | ||
| + | |||
| + | =====Results===== | ||
| + | Of all 4 measurements, the average was taken an is summarized in the chart below. | ||
| + | |||
| + | [[File:T--UZurich--NLuc_Assay_1.png|600px|thumb|none|Figure 1: Average Luminescence Levels over time after 30 minutes incubation]] | ||
| + | |||
| + | As expected the control sample (UT) did not show any luminescence in the presence of the NanoLuc substrate.<br> | ||
| + | eBAK1 coexpressed with eEFR showed a strong increase in luminescence. However, the highest levels were recorded in samples without the bacterial epitope elf18. This was unexpected as the epitope elf18 is the known elicitor to induce ligand-dependent interaction between the EFR receptor and its coreceptor BAK1 in <i>A. thaliana</i>. We later repeated the assay with the EFR ectodomain to get more data. Presence of the bacterial epitope elf18 did not lead to an increase in fluorescence levels again. | ||
| + | |||
| + | eBAK1 coexpressed with eCORE showed an increase in luminescence, although the effect was much smaller when compared with eEFR. The results again suggest, that addition of the bacterial elicitor csp22, which initiates interaciton between CORE and BAK1 does not increase the luminescence levels as samples without csp22 added showed greater luminescence than samples which were treated with this bacterial epitope. | ||
| + | |||
| + | These results suggest that our plasmids get expressed. It further has been shown that the NanoBit parts fused to the receptors are able to interact and reconstitute their functionality as a funcitonal NanoLuc protein which catalyzes the reaction of furimazine to furimamide, which gives a luminescent output. | ||
| + | In our case, however, receptor-specific bacterial epitopes did not increase luminescence levels when the receptors were expressed in <i>S. cerevisiae</i>. | ||
| + | |||
| + | ====Luminescence Assay 2==== | ||
| + | |||
| + | The assay was repeated in the a similar manner with the following differences: | ||
| + | *We did not test again for samples which had been transformed with eCORE plasmids. So we only had samples for untransformed yeast cells and for cells transfected with the plasmids containting [[Part:BBa K3610038]] and [[Part:BBa K3610043]]. | ||
| + | *The optical density of all samples was adjusted to OD600 = 0.26. | ||
| + | *Measurement started immediately after addition of the bacterial elicitor and the NanoGlo solution. | ||
| + | |||
| + | [[File:T--UZurich--NLuc_Assay_2.png|600px|thumb|none|Figure 10: Average Luminescence Levels over time, measured directly after addition of bacterial elicitor and furimazine]] | ||
| + | |||
| + | Our observations of the second luminescence assay matched our expectations after the first one. Luminescence levels were indeed increased when the cells had previously been transformed with our constructs. It was again the case, however, that reconstitution of the NanoLuc protein was not driven by the ligand-dependent interaction of the plant receptors. It seemed, again, to be the case, that addition of bacterial elicitor did in fact decrease the measured luminescent output. | ||
| + | |||
| + | |||
| + | ====Luminescence Assay 3==== | ||
| + | The assay was repeated again with the following differences (compared to the first assay): | ||
| + | *We did not test again for samples which had been transformed with eCORE plasmids. So we only had samples for untransformed yeast cells and for cells transfected with the plasmids containting [[Part:BBa K3610038]] and [[Part:BBa K3610043]]. | ||
| + | *NanoGlo was added before the measurements were made, the bacterial elicitor was added right before measurement | ||
| + | *Optical density was adjusted to OD600 = 0.5. We waited, however, for almost two hours, during which the cells were kept in TE buffer. During this time, a lot of the cells died, which decresaed the amount of whole living cells as TE supports cell lysis. | ||
| + | *Only one bacterial epitope (elf18) was added. | ||
| + | |||
| + | [[File:T--UZurich--NLuc_Assay_3.png|600px|thumb|none|Figure 11: Average Luminescence Levels over time, furimazine added 30 minutes before measurement and bacterial elicitor added immediately before.]] | ||
| + | |||
| + | A third time, we were able to observe that expression of our constructs increased luminescence intensity when the substrate furimazine was added. We, additionally, observed that the increase in luminescence was not initiated or significantly enhanced by presence of the bacterial epitope elf18. | ||
| + | |||
| + | ====Summary of Luminescence Assays==== | ||
| + | We conducted for assays with a plate reader of the type <i>Synergy H1</i>. Each time, luminescence was strongly increased when the samples had been transfected with plant PRRs fused to the NanoBits. This has especially been the case when the plant recpetors EFR and its coreceptor BAK1 were used. | ||
| + | The increase in luminescence intensity did not seem to be driven by epitope.dependent interaciton of the two receptors. It is possible that, unlinke plants, <i>S. cerevisiae</i> cells lack any mechanisms to prevent unprovoked interaction between the target-receptor EFR and BAK1. Another possible explanation for the results could be that the receptors proteins are not processed the same way as they are in plant cells. For example, in plant cells, the receptor proteins get highly glycosylated. It is possible that this glycosylation influences the structure and the functionality of the receptor proteins. | ||
| + | |||
| + | ==Conclusion UZürich 2020== | ||
| + | The BAK1 ectodomain was a crucial part of our project. Nothing we set out to do could have worked without this part. We are happy with what we have achieven, even though there is so much more left to discover, much more to try and apply. | ||
| + | |||
| + | We are nevertheless happy, to have used this part in the following ways: | ||
| + | *Demonstration expression by fusing this part to YFP and transfecting <i>S. cerevisiae</i> cells with the construct. We saw that the cells express the receptor and, in addition, some of it gets trafficked to the plasma membrane. | ||
| + | *Dimerization Assay with eEFR and split-mCherry. Fused to the split-mCherry parts, coexpression of EFR and BAK1 did not reveal increased fluorescence when compared to the control. Interestingly, there seems to be an effect of the bacterial elicitor elf18 on fluorescence intensity. | ||
| + | *Dimerization Assay with EFR, CORE and split-NanoLuc. Fused to the NanoBit parts, coexpression of this part and either target receptor EFR or CORE leads to a luminescent output in the prescence of the NanoLuc substrate furimazine. It does, however, seem that dimerization of the split-NanoLuc is not driven by ligand-dependent interaction of the plant PRRs. | ||
| + | |||
<!-- --> | <!-- --> | ||
Latest revision as of 01:42, 28 October 2020
BAK1 Ectodomain from Arabidopsis thaliana
This part is a version of the BAK1 receptor from A. thaliana (Part:BBa_K3610001 for full length) without the intracellular kinase domain and without the native signal sequence.
Contents
Usage and Biology
The BRI1-associated receptor kinase (BAK1) is a leucine-rich repeat receptor kinase (LRR-RK) which interacts with multiple other LRR-RKs with different functions in hormone signalling and defense response. BAK1 localizes at the plasma membrane and the endosome. The BAK1 protein forms a structure with an extracellular domain with leucine-rich repeats, a single pass transmembrane domain and an intracellular domain with a kinase function.
Among others, BAK1 interacts with the LRR-RKs EF-Tu receptor (EFR), Flagellin sensing 2 (FLS2) and cold-shock protein receptor (CORE), all of which are pathogen recognition receptors (PRR) in brassicaceae plants. Upon binding of a microbe-associated molecular pattern at the LRR domain of the PRR, BAK1 forms a heterodimer with the PRR which triggers a phosphorylation cascade, leading to upregulation of defense mechanisms.
This part includes only the ectodomain and the transmembrane domain. The intracellular kinase domain was cleaved off. Additionally, the natural signal peptide sequence, which is necessary for localization at the cell membrane in the plant, has been cleaved off as well. Therefore, this part needs to be used together with a signal peptide from the organism in which the protein is expressed.
Plant Immunity Based Biosensing UZürich 2020
For visualizing the interaction of BAK1 with other plant receptors the cytoplasmic domain can be replaced with a split fluorescent protein or another protein that generates a visual output. For our iGEM project, we designed a system to use this mechanism for detection of bacterial epitopes in water. We used this part for multiple experiments.
Expression with YFP
We fused this part together with the yellow fluorescent protein venus with a 15 amino acids long linker and added the secretion signal from the alpha-Factor, a protein from S. cerevisiae. The corresponding composite part is Part:BBa_K3610032 (the results for this experiment are also on this parts page). We expressed this protein in S. cerevisiae to test two things. First of all, we wanted to see whether the protein gets expressed at all, and secondly, we were interested in seeing, whether the secretion signal from yeast and the receptor without the intracellular kinase domain would be sufficient for localization at the cell periphery.
In a first step we inserted the single fragments making up this part into a plasmid with a gentamycin-3-acetyltransferase gene and transformed E. coli (DH10alpha) with the plasmids for amplification. In the next step we assembled the fragments in a plasmid with a spectinomycin acetyltransferase and amplified the plasmids again in the same E. coli strain. For this step we applied the techniques of Golden Gate Cloning to get the fragments in the right order into the plasmid. The restriction enzyme we chose was BsaI. For expressing this part consisting of YFP and the receptor protein, we initially intended to use promoters of different strengths to get more quantitative data. Finally, we got the construct in a plasmid with a truncated version of the ADH1 promoter from S. cerevisiae. For termination, this part has the terminator sequence of the enolase 2 protein from S. cerevisiae. The plasmid also contained the TRP1 gene, which encodes phosphoribosylanthranilate isomerase, an enzyme that catalyzes the third step in tryptophan biosynthesis. This enabled us to use the same plasmid for expression in S. cerevisiae. We prepared a medium containing YNB and free amino acids, without tryptophan. S. cerevisiae cells (AP4) were transfected with the plasmid and then plated on the selective medium.
Microscopy
After successful transformation of yeast cells we checked for expression of the protein under a confocal microscope.
Confocal microscopy confirmed increased fluorescence in the S. cerevisiae cells that had been previously transfected with plasmids containing BAK1 ectodomain fused to YFP. This increased fluorescence indicates expression of our genes. As we were particularily interested in seeing where within the cell the receptor would get localized, we stained the membrane with the popular stain FM4-64 and then examined the samples again with confocal microscopy.
This additional imaging experiment revealed that the fluorescent protein is in part localized at the cell periphery. This is in alignment with our expectations as our construct includes a secretion signal protein and a receptor coding protein with the transmembrane domain. These results suggest that the secretion peptide fused to the receptor ectodomain, including the transmembrane domain can be expressed in S. cerevisiae and that the components are sufficient for localization at the cell membrane.
This observation is already exciting and underlines plant PRR's potential as tools for Synthetic Biology.
Spectrometry
In addition to analyzing the cells with a microscope, we conducted a fluorescence assay with a plate reader. We conducted this experiment for multiple receptors at the same time. This way we were able to compare the expression levels of differnt versions of the BAK1 receptor. For each receptor we tried to isolate three different biological samples, however, not all cells grew. Ultimately, we only had two samples for the following S. cerevisiae cells: untransformed (Control), transformed with BAK1 ectodomain fused to YFP (eBAK) and the CORE ectodomain fused to YFP (eCORE). For the BAK1 with and without the native signal peptide fused to YFP (BAK+ and BAK-) and the EFR ectodomain fused to YFP (eEFR), we had samples from three different colonies. For each biological replicate, the optical density at absorbance of 600 nm (OD600) and the fluorescence levels were measured three times.
| measured OD600 values (OD) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Replicate 1 | Replicate 2 | Replicate 3 | |||||||
| Blank | 0,08200000226 | 0,08200000226 | 0,08389999717 | ||||||
| Control | 0,3806000054 | 0,3747999966 | 0,4221999943 | 0,1316999942 | 0,131400004 | 0,1176000014 | |||
| BAK+ | 0,4943000078 | 0,4638999999 | 0,4514000118 | 0,5781000257 | 0,5253999829 | 0,5799999833 | 0,2615999877 | 0,2171999961 | 0,2011999935 |
| BAK- | 1,417099953 | 1,365499973 | 1,368899941 | 0,6305999756 | 0,5633999705 | 0,6216999888 | 0,896600008 | 0,7882999778 | 0,8032000065 |
| eBAK | 1,009699941 | 0,8404999971 | 0,8934999704 | 0,2653000057 | 0,2368000001 | 0,2592999935 | |||
| eCORE | 1,021499991 | 0,8616999984 | 0,9178000093 | 0,826300025 | 0,6888999939 | 0,7401999831 | |||
| eEFR | 1,379699945 | 1,322700024 | 1,333500028 | 1,035899997 | 1,014000058 | 0,9526000023 | 0,4860999882 | 0,3797000051 | 0,3829999864 |
The following settings were applied for fluorescence measurements:
| Mode: | Fluorescence Top Reading |
| Excitation Wavelength: | 485 nm |
| Emission Wavelength: | 535 nm |
| Excitation Bandwidth: | 20 nm |
| Emission Bandwidth: | 25 nm |
| Temperature: | 22.3°C |
| Fluorescence Top Reading (FTR) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Replicate 1 | Replicate 2 | Replicate 3 | |||||||
| Blank | 1297 | 1282 | 1322 | ||||||
| Control | 2684 | 2474 | 2634 | 1852 | 1792 | 1750 | |||
| BAK+ | 3038 | 2813 | 2760 | 2836 | 2493 | 2788 | 2084 | 2072 | 2067 |
| BAK- | 35794 | 30319 | 31424 | 10792 | 9097 | 10517 | 22609 | 20227 | 21220 |
| eBAK | 26455 | 19828 | 21613 | 6614 | 5507 | 6229 | |||
| eCORE | 10709 | 8382 | 9339 | 8957 | 7062 | 7735 | |||
| eEFR | 43125 | 37782 | 39589 | 25641 | 24668 | 22517 | 12410 | 9054 | 9027 |
After measurement of the optical density and the fluorescence, the data were blank corrected (the average of the three blank measurements was subtracted from each measurement value).
The average of each of the three (or two) samples was calculated. From these values, the average was taken again.
After this step, we normalized the fluorescent output for OD600 (FTR/OD). The results of these calculations are displayed in the table below.
| Control | BAK+ | BAK- | eBAK | eCORE | eEFR |
|---|---|---|---|---|---|
| 4185,221063 | 9731,614266 | 26067,19254 | 28118,24739 | 3712,946478 | 23379,84399 |
If we set the values for the Control to 1 (Control = 1), then we get the fluorescence levels relative to the control, which is again diplayed in the table below.
| Control | eCORE | eEFR | BAK- | BAK+ | eBAK |
|---|---|---|---|---|---|
| 1 | 0,8871565975 | 5,586286516 | 6,228390841 | 2,325233033 | 6,718461693 |
Results from the plate reader and from confocal microscopy were in alignment. Measurement with the fluorometer suggested increased expression of YFP when S. cerevisiae cells are transfected with a plasmid that contains the BAK1 ectodomain fused to YFP. These results together strongly imply, that the ectodomain of the plant PRR BAK1 from A. thaliana, which has been fused to the secretion signal from the alpha-Factor, is expressed in yeast and also gets localized at the cell membrane.
Flow Cytometry
It has been important to us to examine a sample with different approaches simultaneously, which is why we were eager to also measure fluorescence intensity by flow cytometry. In a first phase, 100,000 cells were measured from each biological replicate (488/530 FITC channel in a BD FACSCanto II flow cytometer). In the next phase, the biological replicates for each construct were pooled together and 200,000 cells from each sample were measured.
Flow catometry provided further evidence for expression of the construct in S. cerevisiae. Cells transfected with plasmids containing BAK- showed significantly increased fluorescence intensities when examined as single biological replicates, as well as when the replicates were pooled together in one sample.
Expression with split-mCherry
For another part of our project, we fused the ectodomain to the N-terminal domain of the split-mCherry protein instead of YFP. The goal was to coexpress this construct (Part:BBa_K3610034) with one of the plant PRRs that show ligand-dependent interaction with BAK1, like EFR. We managed to design a construct consisting of the ectodomain of EFR which was fused to a secretion signal and the C-terminal domain of mCherry (Part:BBa_K3610039. Coexpressed in S. cerevisiae, these two parts are able to interact upon ligand-binding. The goal of our experiments was to test, whether ligand-dependent dimerization of the mCherry protein would be sufficient to drive dimerization of the split-mCherry parts.
During our project, we achieved coexpression of Part:BBa_K3610034 with Part:BBa_K3610042, which encodes the ectodomain of the plant PRR target receptor EFR fused to the C-terminal domain of mCherry in S. cerevisiae.
Fluorescence Assay
To test, whether the EFR ligand elf18 would drive dimerization of the receptors, we conducted an experiment, during which we compared the fluorescent response of the yeast.
We therefore conducted an assay with a fluorometer, a plate reader of the type Synergy H1 (λEX = 587nm and λEM = 610nm). We eamined untransformed cells as a control (UT) and cells which had been previously transfected with plasmids containting the parts Part:BBa_K3610034 and Part:BBa_K3610042 (eEFR). All samples had been adjusted to OD600 = o0.5 before the plate reader experiment.
The following samples were prepared:
- UT
- UT, elf18 added
- eEFR
- eEFR, elf18 added
Surprisingly, fluorescence levels did increase when the bacterial elicitor was present, however, this was the case for both types of samples. We must be, however, aware of the fact that the variance was rather large and this one time assay does not provide enough data to confirm a significant effect of the bacterial elicitor. What was an even bigger surprise is that the measured fluorescence intensities were higher for the samples containing the untransformed yeast cells, both when the bacterial elicitor was present and when it had not been added. The reasons for these results are unclear. It could be due to an error when adjusting the samples to the same OD600. A difference in the OD could lead to increased fluorescence levels. This would still not explain why the bacterial elicitor seemed to increase autofluorescence for the untransformed yeast cells.
It is clear that this deserves further examination. We propose to repeat the measurements to increase the data coverage and rule out errors during pipetting or labelling the samples. It should further be attempted to lower autofluorescence for yeast cells that resembles the fluorescence of mCherry. It further needs to examined, whether the same type of results are obtained when a different split-reporter is fused to the receptor domain (something which we were able to carry out during our project).
Expression with split-NanoLuc
For another part of our project, we fused the ectodomain to the LargeBit part of the split-NanoLuc protein instead of the intracellular kinase domain. The goal was to coexpress this construct (Part:BBa_K3610032) with one of the plant PRRs that show ligand-dependent interaction with BAK1, for example EFR. We managed to design a construct consisting of the ectodomain of EFR which was fused to a secretion signal and the SmallBit of NanoLuc (Part:BBa_K3610043. Coexpressed in S. cerevisiae, these two parts are able to interact upon ligand-binding. This dimerization will reconstitute the functionality of the NanoLuc protein, leading to a chemiluminescent output in the presence of furimazine.
Luminescence Assay 1
We managed to coexpress the BAK1 ectodomain with LargeBit together with two different receptor ectodomains fused to the SmallBit part of NanoLuc, EFR (eEFR) and CORE (eCORE).
After transfection of S. cerevisiae cells, samples were prepared and examined with a luminometer. We used a plate reader of the type Synergy H1. For a more detailed view on data visite the page of the composite part: Part:BBa K3610038.
Samples were prepared:
- untransfected control (UT)
- eEFR and eBAK1 (eEFR)
- eCORE and eBAk1 (CORE)
each sample type was used for three types of measurements:
- no bacterial elicitor added
- csp22 added
- elf18 added
Applying all setups to each sample, we measured luminescence intensities for 9 different groups.
Results
Of all 4 measurements, the average was taken an is summarized in the chart below.
As expected the control sample (UT) did not show any luminescence in the presence of the NanoLuc substrate.
eBAK1 coexpressed with eEFR showed a strong increase in luminescence. However, the highest levels were recorded in samples without the bacterial epitope elf18. This was unexpected as the epitope elf18 is the known elicitor to induce ligand-dependent interaction between the EFR receptor and its coreceptor BAK1 in A. thaliana. We later repeated the assay with the EFR ectodomain to get more data. Presence of the bacterial epitope elf18 did not lead to an increase in fluorescence levels again.
eBAK1 coexpressed with eCORE showed an increase in luminescence, although the effect was much smaller when compared with eEFR. The results again suggest, that addition of the bacterial elicitor csp22, which initiates interaciton between CORE and BAK1 does not increase the luminescence levels as samples without csp22 added showed greater luminescence than samples which were treated with this bacterial epitope.
These results suggest that our plasmids get expressed. It further has been shown that the NanoBit parts fused to the receptors are able to interact and reconstitute their functionality as a funcitonal NanoLuc protein which catalyzes the reaction of furimazine to furimamide, which gives a luminescent output. In our case, however, receptor-specific bacterial epitopes did not increase luminescence levels when the receptors were expressed in S. cerevisiae.
Luminescence Assay 2
The assay was repeated in the a similar manner with the following differences:
- We did not test again for samples which had been transformed with eCORE plasmids. So we only had samples for untransformed yeast cells and for cells transfected with the plasmids containting Part:BBa K3610038 and Part:BBa K3610043.
- The optical density of all samples was adjusted to OD600 = 0.26.
- Measurement started immediately after addition of the bacterial elicitor and the NanoGlo solution.
Our observations of the second luminescence assay matched our expectations after the first one. Luminescence levels were indeed increased when the cells had previously been transformed with our constructs. It was again the case, however, that reconstitution of the NanoLuc protein was not driven by the ligand-dependent interaction of the plant receptors. It seemed, again, to be the case, that addition of bacterial elicitor did in fact decrease the measured luminescent output.
Luminescence Assay 3
The assay was repeated again with the following differences (compared to the first assay):
- We did not test again for samples which had been transformed with eCORE plasmids. So we only had samples for untransformed yeast cells and for cells transfected with the plasmids containting Part:BBa K3610038 and Part:BBa K3610043.
- NanoGlo was added before the measurements were made, the bacterial elicitor was added right before measurement
- Optical density was adjusted to OD600 = 0.5. We waited, however, for almost two hours, during which the cells were kept in TE buffer. During this time, a lot of the cells died, which decresaed the amount of whole living cells as TE supports cell lysis.
- Only one bacterial epitope (elf18) was added.
A third time, we were able to observe that expression of our constructs increased luminescence intensity when the substrate furimazine was added. We, additionally, observed that the increase in luminescence was not initiated or significantly enhanced by presence of the bacterial epitope elf18.
Summary of Luminescence Assays
We conducted for assays with a plate reader of the type Synergy H1. Each time, luminescence was strongly increased when the samples had been transfected with plant PRRs fused to the NanoBits. This has especially been the case when the plant recpetors EFR and its coreceptor BAK1 were used. The increase in luminescence intensity did not seem to be driven by epitope.dependent interaciton of the two receptors. It is possible that, unlinke plants, S. cerevisiae cells lack any mechanisms to prevent unprovoked interaction between the target-receptor EFR and BAK1. Another possible explanation for the results could be that the receptors proteins are not processed the same way as they are in plant cells. For example, in plant cells, the receptor proteins get highly glycosylated. It is possible that this glycosylation influences the structure and the functionality of the receptor proteins.
Conclusion UZürich 2020
The BAK1 ectodomain was a crucial part of our project. Nothing we set out to do could have worked without this part. We are happy with what we have achieven, even though there is so much more left to discover, much more to try and apply.
We are nevertheless happy, to have used this part in the following ways:
- Demonstration expression by fusing this part to YFP and transfecting S. cerevisiae cells with the construct. We saw that the cells express the receptor and, in addition, some of it gets trafficked to the plasma membrane.
- Dimerization Assay with eEFR and split-mCherry. Fused to the split-mCherry parts, coexpression of EFR and BAK1 did not reveal increased fluorescence when compared to the control. Interestingly, there seems to be an effect of the bacterial elicitor elf18 on fluorescence intensity.
- Dimerization Assay with EFR, CORE and split-NanoLuc. Fused to the NanoBit parts, coexpression of this part and either target receptor EFR or CORE leads to a luminescent output in the prescence of the NanoLuc substrate furimazine. It does, however, seem that dimerization of the split-NanoLuc is not driven by ligand-dependent interaction of the plant PRRs.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 576
Illegal PstI site found at 621 - 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 576
Illegal PstI site found at 621 - 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 576
Illegal PstI site found at 621 - 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 576
Illegal PstI site found at 621 - 1000COMPATIBLE WITH RFC[1000]