Part:BBa_K3610001
BAK1 from Arabidopsis thaliana
This part contains the cDNA of the full-length plant cell surface receptor BAK1 from A. thaliana.
Usage and Biology
The BRI1-associated receptor kinase (BAK1) is a leucin-rich repeat receptor kinase (LRR-RK) which interacts with multiple other LRR-RKs with different functions in hormone signalling and defense response. BAK1 localizes at the plasma membrane and the endosome. The BAK1 protein forms a structure with an extracellular domain with leucin-rich repeats, a single pass transmembrane domain and an intracellular domain with a kinase function.
Among others, BAK1 interacts with the LRR-RKs EF-Tu receptor (EFR), Flagellin sensing 2 (FLS2) and cold-shock protein receptor (CORE), all of which are pathogen recognition receptors (PRR) in brassicaceae plants. Upon binding of a microbe-associated molecular pattern at the LRR domain of the PRR, BAK1 forms a heterodimer with the PRR which triggers a phosphorylation cascade, leading to upregulation of defense mechanisms.
Characterization
In our project we used this receptor to demonstrate expression of plant PRRs in S. cerevisiae by expressing this part with a fluorescent marker fused to the N-terminal end of the sequence.
For a more detailed characterization of these experiments part see Part:BBa_K3610030, where all experiments are described more acurately.
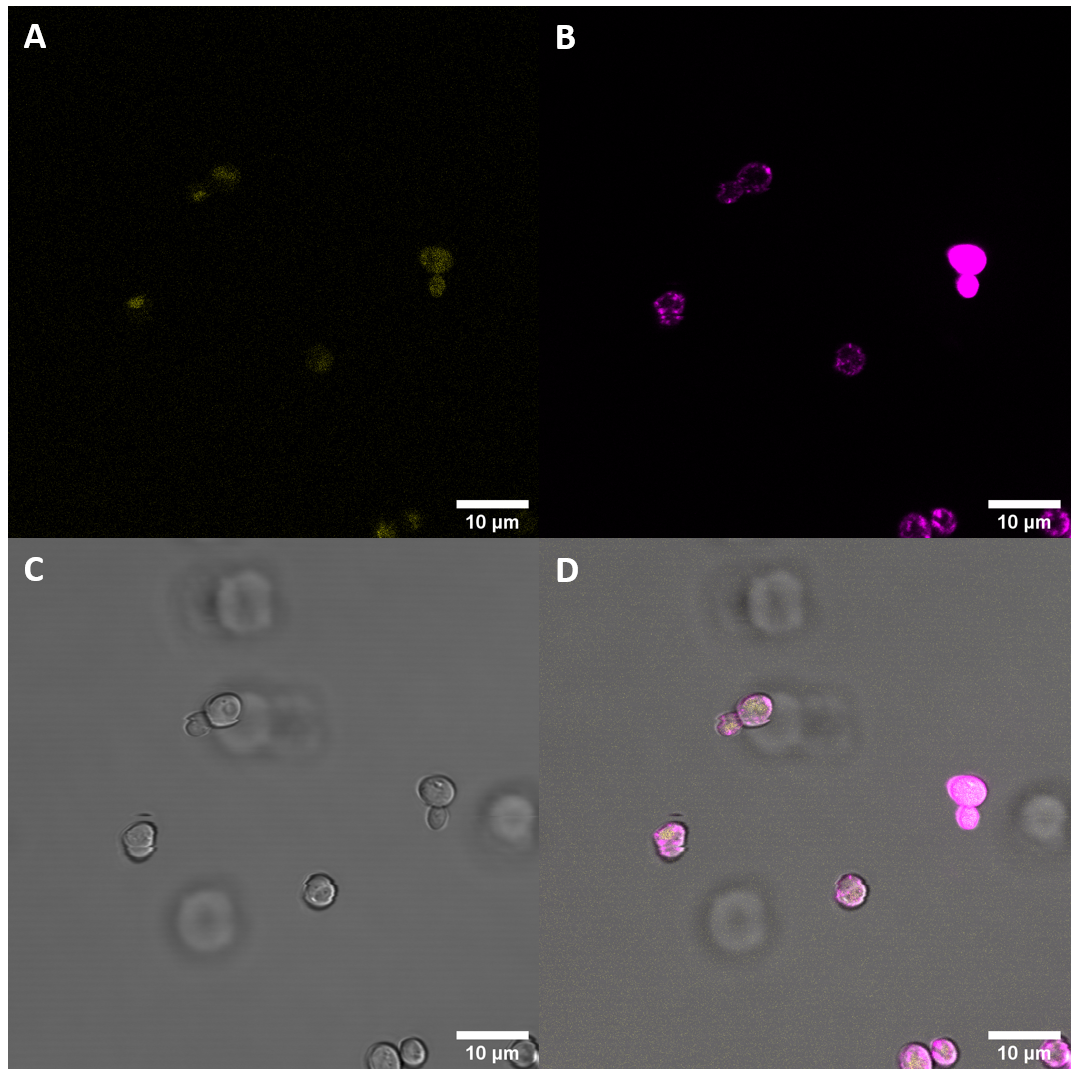
Fluorescence Microscopy
We expressed this part fused to YFP in S. cerevisiae and then examined the cells with meand of confocal fluorescence microscopy. We also stained the membrane to see if the protein would localize at the cell periphery.
Fluorescence Microscopy suggests that there is some expression of the BAK+ construct. It fails, however, to get localized at the cell periphery. Nevertheless, expression of the full length receptor in S. cerevisiae can already be seen as a considerable success.
Plate Reader
In addition to analyzing the cells with a microscope, we conducted a fluorescence assay with a plate reader. We conducted this experiment for multiple receptors at the same time. This way we were able to compare the expression levels of differnt versions of the BAK1 receptor. The cosntruct containing this part is referred to as BAK+.
Measurements with the luminometer further suggested that BAK+ gets expressed, although it may be expressed at lower levels when compared with other receptors.
Flow Cytometry
In a first phase, 100,000 cells were measured from each biological replicate (488/530 FITC channel in a BD FACSCanto II flow cytometer). In the next phase, the biological replicates for each construct were pooled together and 200,000 cells from each sample were measured.
Flow catometry provided further evidence for expression of the receptor.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal SpeI site found at 1560
Illegal PstI site found at 650
Illegal PstI site found at 695
Illegal PstI site found at 985 - 12INCOMPATIBLE WITH RFC[12]Illegal SpeI site found at 1560
Illegal PstI site found at 650
Illegal PstI site found at 695
Illegal PstI site found at 985 - 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal SpeI site found at 1560
Illegal PstI site found at 650
Illegal PstI site found at 695
Illegal PstI site found at 985 - 25INCOMPATIBLE WITH RFC[25]Illegal SpeI site found at 1560
Illegal PstI site found at 650
Illegal PstI site found at 695
Illegal PstI site found at 985 - 1000COMPATIBLE WITH RFC[1000]
| None |