Difference between revisions of "Part:BBa K115001"
Bavandenberg (Talk | contribs) |
(→Functional Parameters: Austin_UTexas) |
||
| (10 intermediate revisions by 3 users not shown) | |||
| Line 16: | Line 16: | ||
| − | + | ||
| − | === | + | |
| − | + | ||
| + | |||
| + | |||
| + | |||
| + | ===3D Structure=== | ||
| + | (The following information is the contribution of SVCE_Chennai 2016) | ||
| + | |||
| + | |||
| + | |||
| + | |||
<!-- --> | <!-- --> | ||
| + | <span class='h3bb'><strong>3D Model when not ligated to any protein</strong></span> | ||
| + | |||
| + | [[Image:RNAtfinal6.gif]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!-- --> | ||
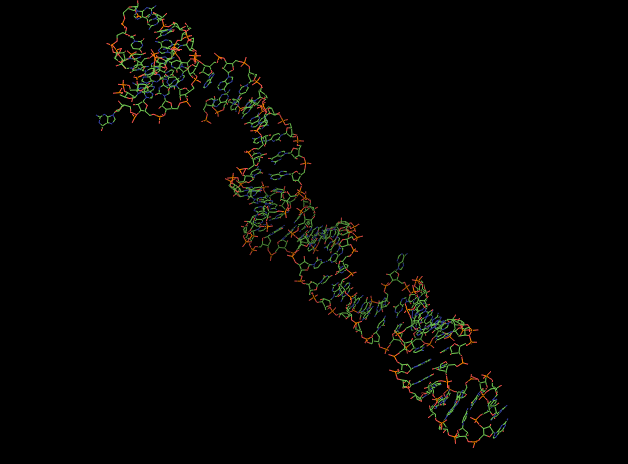
| + | <span class='h3bb'><strong>Another representation of the 3D structure.</strong></span> | ||
| + | |||
| + | [[Image:RNAt2.png]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!--Rice iGEM 2019 improvement --> | ||
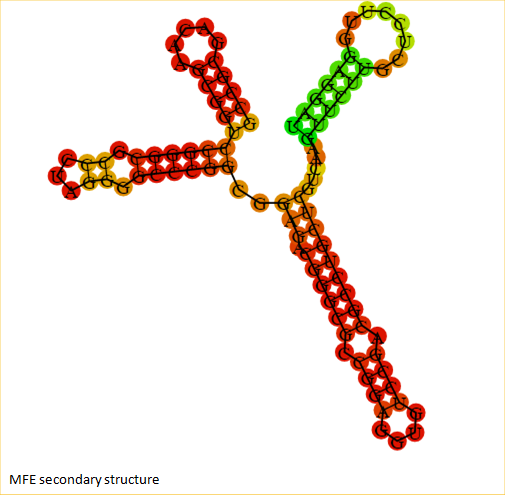
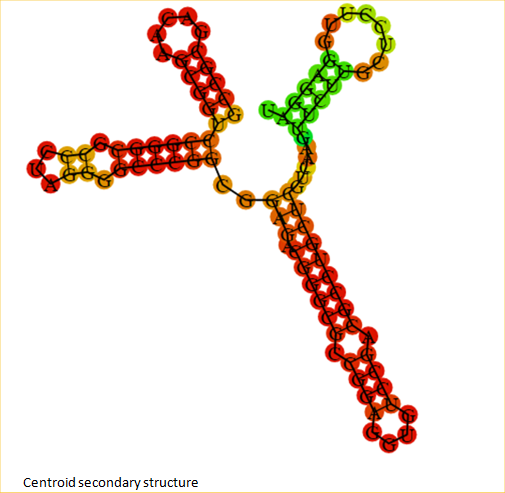
| + | <span class='h3bb'><strong>Secondary Structure when RNA thermometer is not ligated to any protein</strong></span> | ||
| + | |||
| + | [[Image:rnat.png]] | ||
| + | |||
| + | [[Image:rnat2.png]] | ||
| + | |||
| + | ===Characterization=== | ||
| + | <html> | ||
| + | <p>To characterize this RNA thermometer in vivo, we made an RNA thermometer testing circuit, transformed it into <i>E. coli</i>, and tested the fluorescent output at different temperatures. These graphs show the relative fluorescent intensity of each thermometer between either 25 °C and 30 °C or 25 °C and 37 °C</p> | ||
| + | <img src="https://2019.igem.org/wiki/images/a/a2/T--Rice--RNATcassette.svg" width = 60%> | ||
| + | <p><b>Figure 1.</b>RNA thermometer testing circuit</p> | ||
| + | <img src="https://2019.igem.org/wiki/images/1/16/T--Rice--BBa115001.png" width = 60%> | ||
| + | <p><b>Figure 2.</b> Fluorescence intensity at 25 °C, 30 °C, and 37 °C.</p> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==Functional Parameters: Austin_UTexas== | ||
| + | <html> | ||
| + | <body> | ||
| + | <partinfo>BBa_K115001 parameters</partinfo> | ||
| + | <h3><center>Burden Imposed by this Part:</center></h3> | ||
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/f/fa/T--Austin_Utexas--no_burden_icon.png" style = "width:160px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: 3.8 ± 7.2% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | <hr> | ||
| + | |||
| + | |||
| + | |||
| + | ===References=== | ||
| + | |||
| + | <br>SimRNAweb: a web server for RNA 3D structure modeling with optional restraints. | ||
Latest revision as of 19:29, 3 September 2020
RNA thermometer (ROSE)
An [http://2008.igem.org/Team:TUDelft/Temperature_analysis#RNA_thermometer RNA thermometer] that can be used for temperature sensitive post-transcriptional regulation which is based on a RNA thermometer from Bradyrhizobium japonicum that initiates translation at 42°C.
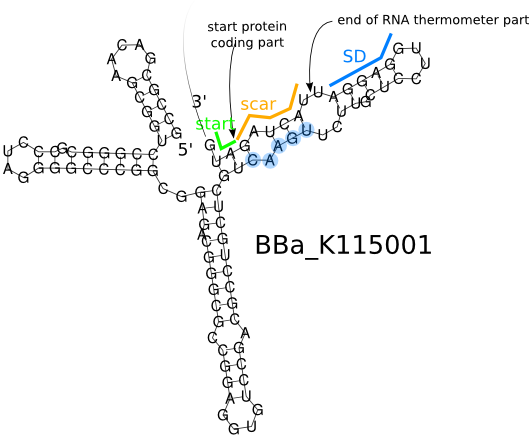
The image shows the secondary structure of the part after ligation to a protein coding part, as predicted by [http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi RNAfold]. Notice that the 3' end, including the scar and the start codon, does not belong to the part. The blue region is the Shine Dalgarno sequence which is the ribosome binding site. The blue nucleotides are the ones that had to be changed in order to get the desired secondary structure after introduction of the scar.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
3D Structure
(The following information is the contribution of SVCE_Chennai 2016)
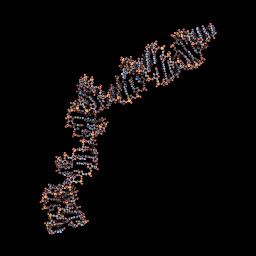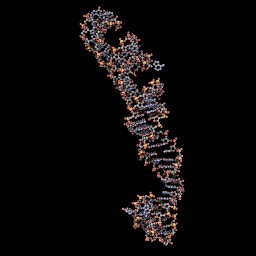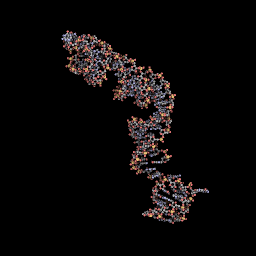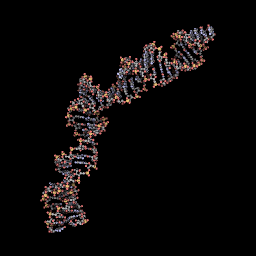
3D Model when not ligated to any protein
Another representation of the 3D structure.
Secondary Structure when RNA thermometer is not ligated to any protein
Characterization
To characterize this RNA thermometer in vivo, we made an RNA thermometer testing circuit, transformed it into E. coli, and tested the fluorescent output at different temperatures. These graphs show the relative fluorescent intensity of each thermometer between either 25 °C and 30 °C or 25 °C and 37 °C

Figure 1.RNA thermometer testing circuit

Figure 2. Fluorescence intensity at 25 °C, 30 °C, and 37 °C.
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
References
SimRNAweb: a web server for RNA 3D structure modeling with optional restraints.