Difference between revisions of "Part:BBa K2926090"
| Line 35: | Line 35: | ||
<div class="quarter right"> | <div class="quarter right"> | ||
<figure class="figure large"> | <figure class="figure large"> | ||
| − | <img class="figure image" src="https://2019.igem.org/wiki/images/6/6d/T--Bielefeld-CeBiTec--Hygromycin_Bl.png"> | + | <img style="width:400px" class="figure image" src="https://2019.igem.org/wiki/images/6/6d/T--Bielefeld-CeBiTec--Hygromycin_Bl.png"> |
<figcaption>Chemical structure of hygromycin B | <figcaption>Chemical structure of hygromycin B | ||
</figcaption> | </figcaption> | ||
| Line 67: | Line 67: | ||
<div class="half left"> | <div class="half left"> | ||
<figure class="figure large"> | <figure class="figure large"> | ||
| − | <img class="figure image" src="https://2019.igem.org/wiki/images/4/47/T--Bielefeld-CeBiTec--basicpart_Intein3d.png" alt=""> | + | <img style="width:400px" class="figure image" src="https://2019.igem.org/wiki/images/4/47/T--Bielefeld-CeBiTec--basicpart_Intein3d.png" alt=""> |
<figcaption>sNMR solution structure of NpuDnaE (PDB code: 2KEQ) (Oeemig et al. 2009)</figcaption> | <figcaption>sNMR solution structure of NpuDnaE (PDB code: 2KEQ) (Oeemig et al. 2009)</figcaption> | ||
</figure> | </figure> | ||
| Line 93: | Line 93: | ||
<div class="middle"> | <div class="middle"> | ||
<figure class="figure large"> | <figure class="figure large"> | ||
| − | <img class="figure image" src="https://2019.igem.org/wiki/images/d/da/T--Bielefeld-CeBiTec--Marker_Intein_Schema.png"> | + | <img style="width:400px" class="figure image" src="https://2019.igem.org/wiki/images/d/da/T--Bielefeld-CeBiTec--Marker_Intein_Schema.png"> |
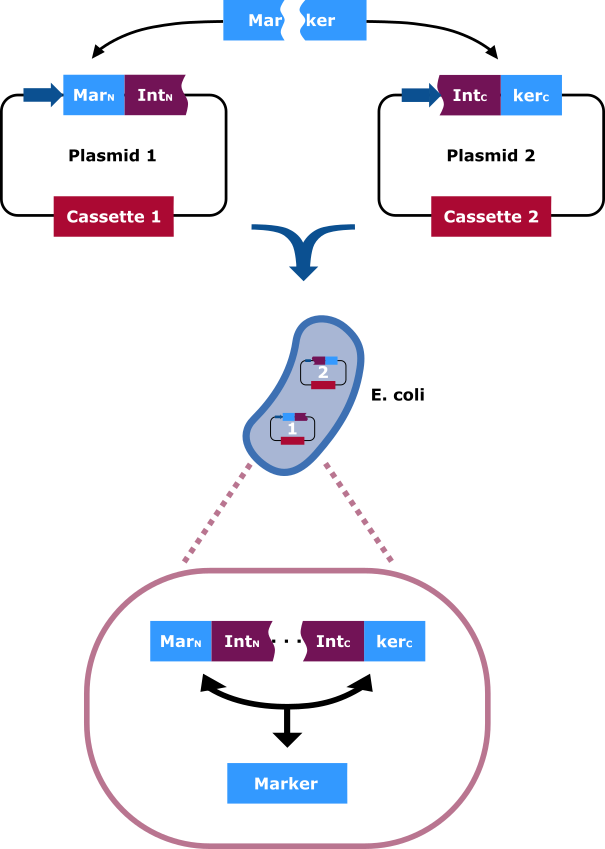
<figcaption>The selection marker is split in two halfs. The N-terminal and C-terminal fragment if it is fused upstream and downstream respectively of the N-terminal and C-terminal fragment of the Npu DnaE intein. Each fusion is cloned in a different backbone, so that the oris are compatible. On the protein level the split intein fragments find to each other and through protein trans-splicing they cut themselves out. Thereby an active hygromycin-B 4-O-kinase is assembled and the cells can grow on hygromycin B rich medium. | <figcaption>The selection marker is split in two halfs. The N-terminal and C-terminal fragment if it is fused upstream and downstream respectively of the N-terminal and C-terminal fragment of the Npu DnaE intein. Each fusion is cloned in a different backbone, so that the oris are compatible. On the protein level the split intein fragments find to each other and through protein trans-splicing they cut themselves out. Thereby an active hygromycin-B 4-O-kinase is assembled and the cells can grow on hygromycin B rich medium. | ||
</figcaption> | </figcaption> | ||
| Line 110: | Line 110: | ||
<div class="half left"> | <div class="half left"> | ||
<figure class="figure large"> | <figure class="figure large"> | ||
| − | <img class="figure image" src="https://2019.igem.org/wiki/images/a/a1/T--Bielefeld-CeBiTec--Plasmide_Hyg_Exteine1.png"> | + | <img style="width:400px" class="figure image" src="https://2019.igem.org/wiki/images/a/a1/T--Bielefeld-CeBiTec--Plasmide_Hyg_Exteine1.png"> |
<figcaption>The fusion on the first plasmid is illustrated. The N-terminal part of the HygR (HygRN) is shown in blue. The N-extein follows with the amino acids tryptophan, leucine and alanine (W L A) on positions -3, -2 and -1, respectively upstream of the N-terminal intein. | <figcaption>The fusion on the first plasmid is illustrated. The N-terminal part of the HygR (HygRN) is shown in blue. The N-extein follows with the amino acids tryptophan, leucine and alanine (W L A) on positions -3, -2 and -1, respectively upstream of the N-terminal intein. | ||
</figcaption> | </figcaption> | ||
| Line 118: | Line 118: | ||
<div class="half right"> | <div class="half right"> | ||
<figure class="figure large"> | <figure class="figure large"> | ||
| − | <img class="figure image" src="https://2019.igem.org/wiki/images/8/82/T--Bielefeld-CeBiTec--Plasmide_Hyg_Exteine2.png"> | + | <img style="width:400px" class="figure image" src="https://2019.igem.org/wiki/images/8/82/T--Bielefeld-CeBiTec--Plasmide_Hyg_Exteine2.png"> |
<figcaption>The fusion on the second plasmid is illustrated. The C-terminal part of the intein is shown. The C-extein follows with the amino acids cysteine, methionine and glutamic acids (C M E) on positions +1, +2 and +3, respectively upstream of the C-terminal HygR (HygRC). | <figcaption>The fusion on the second plasmid is illustrated. The C-terminal part of the intein is shown. The C-extein follows with the amino acids cysteine, methionine and glutamic acids (C M E) on positions +1, +2 and +3, respectively upstream of the C-terminal HygR (HygRC). | ||
</figcaption> | </figcaption> | ||
| Line 129: | Line 129: | ||
<div class="third left"> | <div class="third left"> | ||
<figure class="figure large"> | <figure class="figure large"> | ||
| − | <img class="figure image" src="https://2019.igem.org/wiki/images/4/44/T--Bielefeld-CeBiTec--Single_Hyg.png"> | + | <img style="width:400px" class="figure image" src="https://2019.igem.org/wiki/images/4/44/T--Bielefeld-CeBiTec--Single_Hyg.png"> |
<figcaption>Plate with a hygromycin B concentration of 50 μg/mL. On the upper half, cells were plated that contain only the pSB1Ha3 plasmid. On the lower half, cells were plated that contain only the pSB3Hb3 plasmid. | <figcaption>Plate with a hygromycin B concentration of 50 μg/mL. On the upper half, cells were plated that contain only the pSB1Ha3 plasmid. On the lower half, cells were plated that contain only the pSB3Hb3 plasmid. | ||
</figcaption> | </figcaption> | ||
| Line 149: | Line 149: | ||
<div class="half right"> | <div class="half right"> | ||
<figure class="figure large"> | <figure class="figure large"> | ||
| − | <img class="figure image" src="https://2019.igem.org/wiki/images/4/4f/T--Bielefeld-CeBiTec--Split_Hyg.png"> | + | <img style="width:400px" class="figure image" src="https://2019.igem.org/wiki/images/4/4f/T--Bielefeld-CeBiTec--Split_Hyg.png"> |
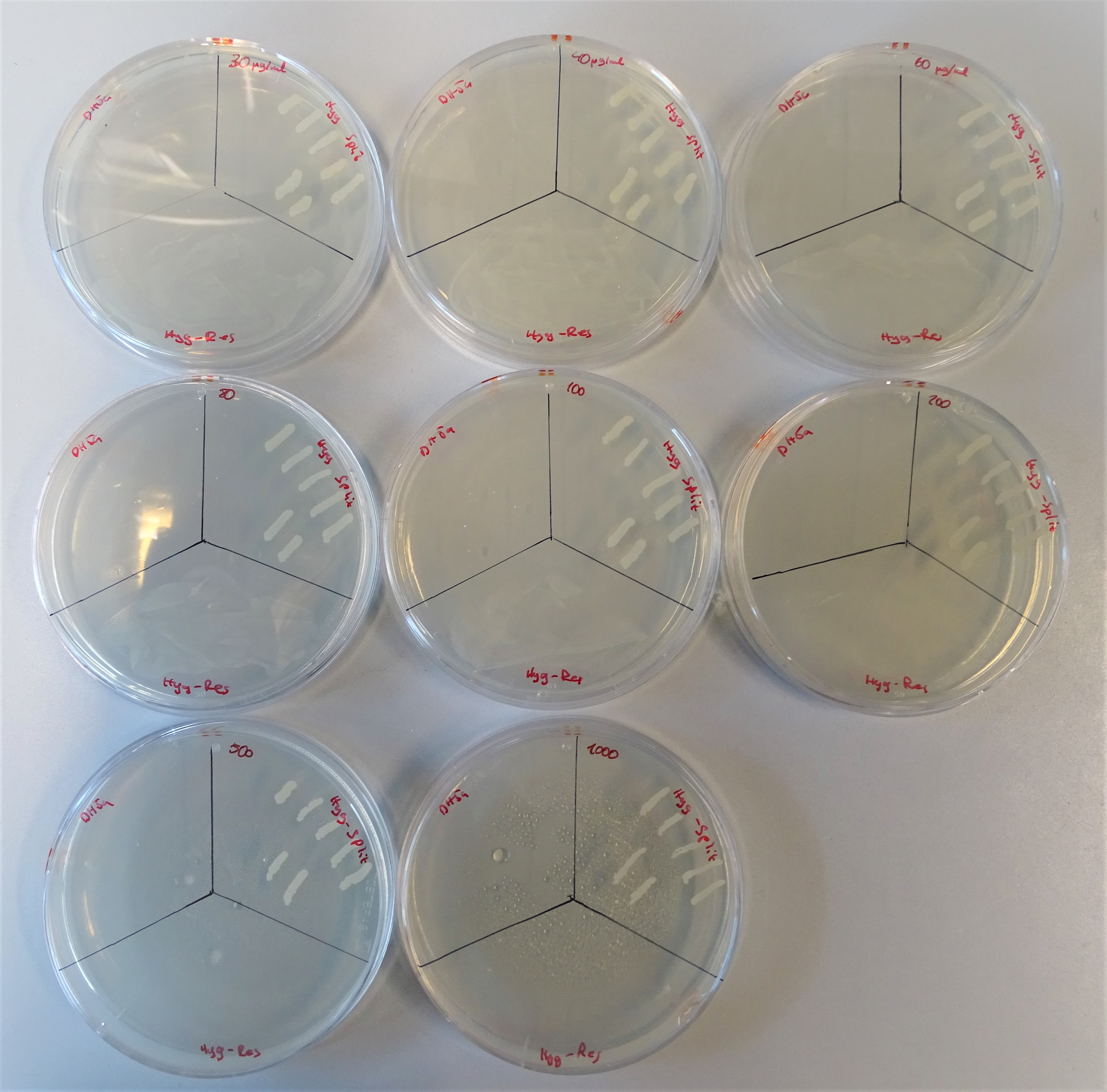
<figcaption>From the upper left site to, to the right till the lower right site, plates are lined up with the hygromycin concentrations 40, 30, 60 80, 100, 200, 500, 1000μg/mL. Every plate is divided into three sections. On the upper left site DH5α was plated representing the negative control, on the upper right site colonies containing both plasmids and on the lower site a colony containing one plasmid with a hygromycin resistance gene. | <figcaption>From the upper left site to, to the right till the lower right site, plates are lined up with the hygromycin concentrations 40, 30, 60 80, 100, 200, 500, 1000μg/mL. Every plate is divided into three sections. On the upper left site DH5α was plated representing the negative control, on the upper right site colonies containing both plasmids and on the lower site a colony containing one plasmid with a hygromycin resistance gene. | ||
</figcaption> | </figcaption> | ||
Latest revision as of 03:51, 22 October 2019
Hygromycin resistance protein Part 2
ghgd
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
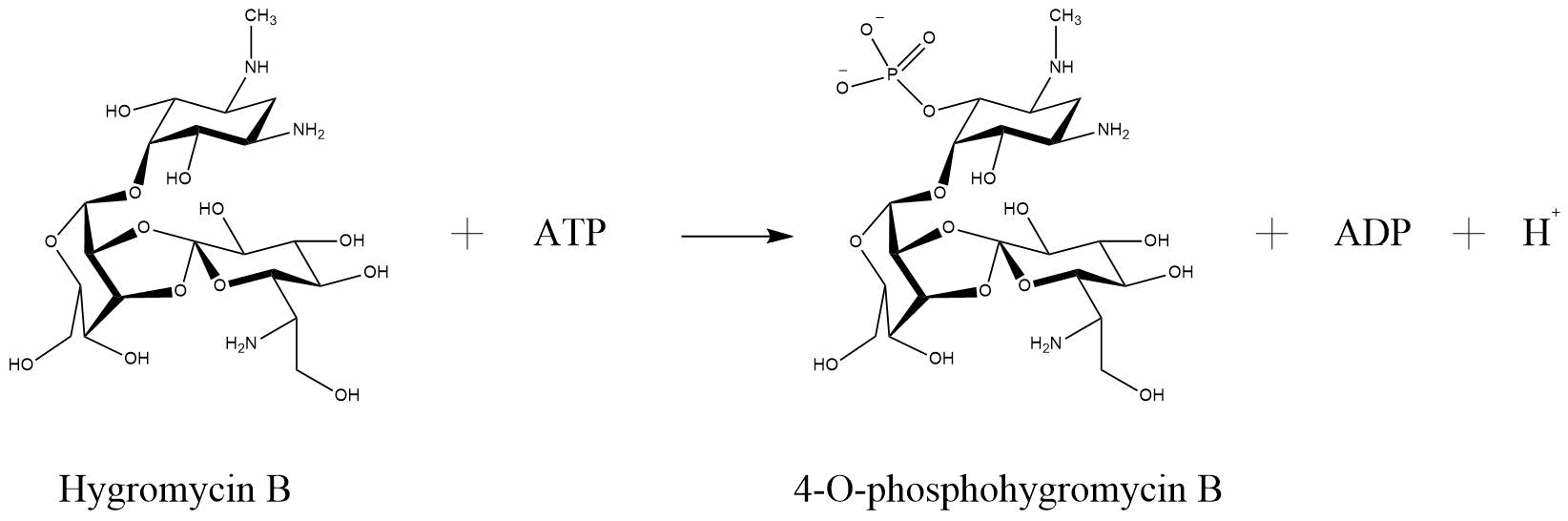
Nowadays it is common for two plasmid systems to be used in the laboratory. In this case it is needed to work with at least two antibiotics to keep the plasmids into the cell. However, it is known, that antimicrobial resistance is becoming a growing threat, as a continued rise in resistance by 2050 would lead to 10 million people dying every year (AMR Review Paper—Tackling a crisis for the health and wealth of nations_1, 2014) and the use of high levels of antibiotics is cost intensive. What if we had an alternative on how to bypass working with two antibiotics? To enable this, we splitted the antibiotic resistance gene for chloramphenicol acetyl transferase (CAT) and cloned each part into a different vector. After co transforming cells with both vectors, through intein-mediated trans-splicing active CAT was produced.
Background
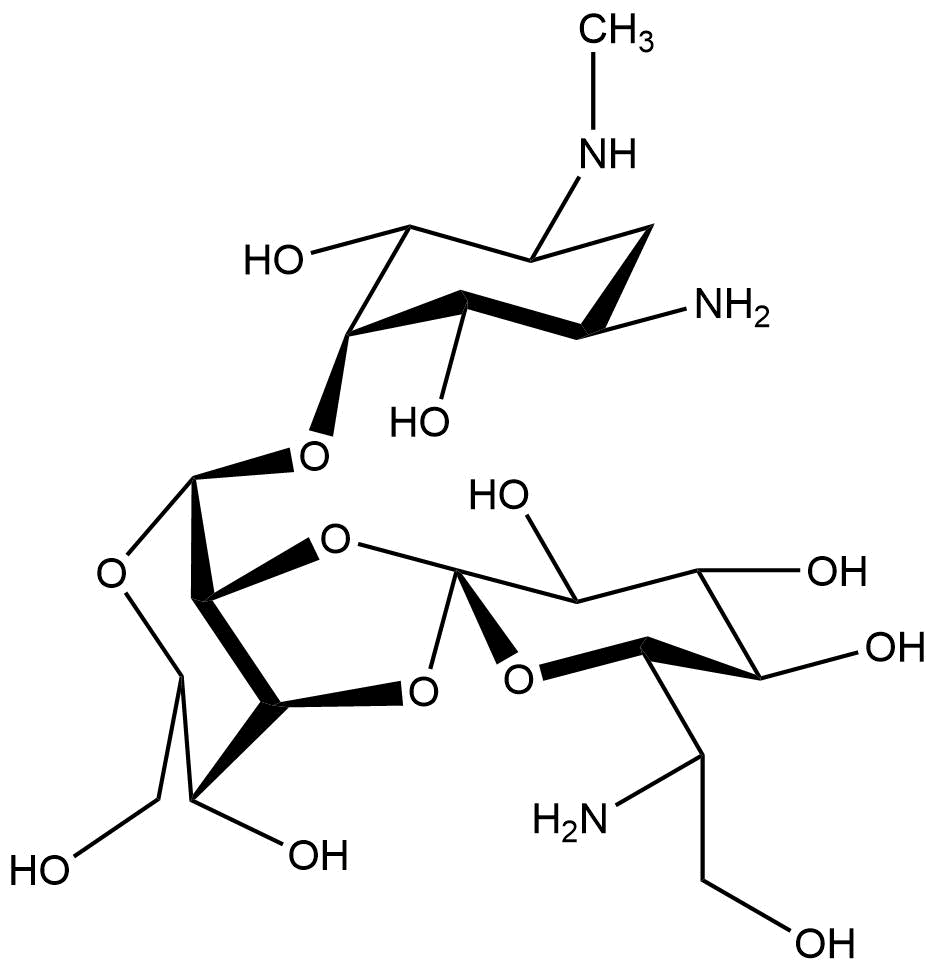
Hygromycin B
Resistance
Npu DnaE Intein

Improved Part
Our aim was to construct two plasmids that express the hygromycin B resistance gene, only by the presence of both plasmids into one E. coli cell. To achieve this, we had to use intein-mediated trans-splicing and choose the optimal split point. Latest, we did orientated on the paper of Jillette, Du, & Cheng, 2018, taking into consideration the amino acids relevant for the splicing process. The information about the flanking sequences the Npu DnaE intein prefers we took from Cheriyan et al., 2013. For hygromycin we chose the split point 240-241 on the amino acid raster.
Mechanism
Plasmid structure


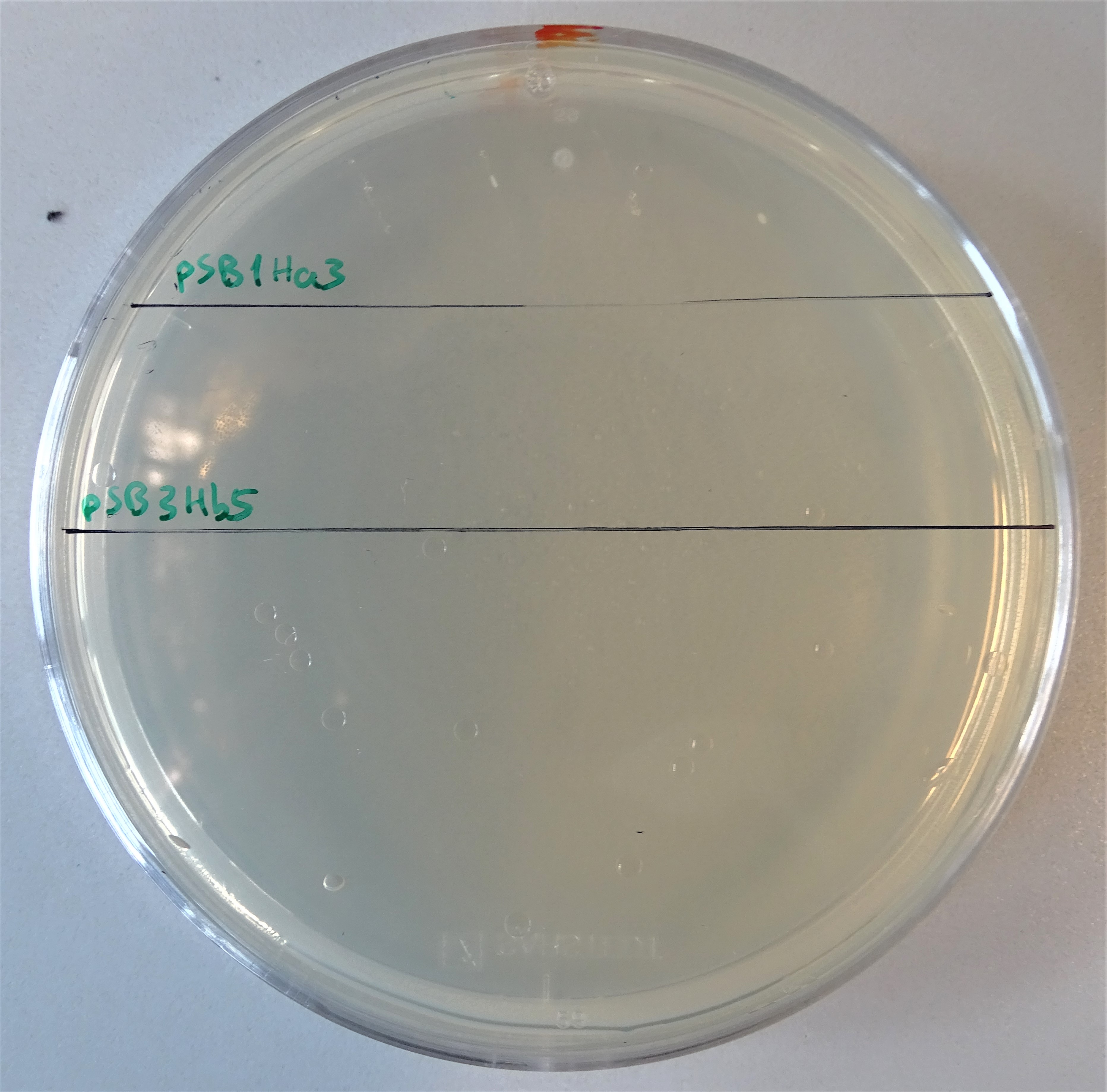
Single plasmid control
After assembling the two plasmids required for expressing an active hph, we plated some kolonies on hygromycin B plates. On the one hand colonies containing the plasmid pSB1Ha3 and on the other hand pSB3Hb5. Through this way we wanted prove, that the half resistance gene fused to an intein is not able to to grow on hygromycin B (Figure 7). Normal working concentrations for hygromycin B is between 200 and 500 µg/mL (Hygromycin B, Thermofisher). Since no colonies grew on the plate it is proven, that cells that contain only one plasmid do not grow even in low hygromycin B concentrations.Growth control