Difference between revisions of "Part:BBa K2924020"
| (16 intermediate revisions by the same user not shown) | |||
| Line 6: | Line 6: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | [[File:AmilCP_Colonies.png|thumb|right|350px|<i><b>Fig.1:</b> LB-Agar plate of Escherichia coli Top10F with the pBbB6c + amilCP plasmid. The amilCP makes the colonies blue.</i>]] | ||
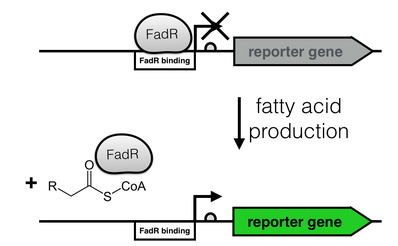
| + | [[File:FadR_mechanism.png|thumb|right|350px|<i><b>Fig. 2</b>: Regulation of the fatty acid metabolism by the transcriptional factor FadR. FadR recognizes its cognate binding site (white), thereby repressing transcription. Upon binding of an Acyl-CoA to FadR, the promoter region is freed, enabling gene expression.</i>]] | ||
<html> <p align="justify"> | <html> <p align="justify"> | ||
| − | + | The blue chromoprotein amilCP from <i>Acropora millepora</i> (<a href="https://parts.igem.org/Part:BBa_K592009">BBa_K592009</a>)<sup>1</sup> was used as a reporter gene for a biosensor. The plan was to implement a chromoprotein as a reporter gene, thus enabling to every laboratory to measure a biosensor without fluorescence applications - in contrast, most laboratories have access to a photometer. The chromoprotein amilCP can be observed with the naked eye, in addition to the measurement method by absorption at the wavelength 588 nm for example with a plate reader. Therefore, this option is available for everyone. | |
</html> | </html> | ||
| + | |||
| + | <html><p align="justify"></html> | ||
| + | The production of long-chain fatty acids (LCFA) is regulated by the transcription factor FadR. This transcription factor can bind to Acyl-CoA which leads to the release of FadR from its cognate operator sequence, thereby increasing gene expression as shown in Fig. 2. | ||
| + | |||
| + | |||
| + | |||
| + | |||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
| − | <partinfo>BBa_K2924020</partinfo> | + | <partinfo>BBa_K2924020 SequenceAndFeatures</partinfo> |
| − | |||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
| Line 21: | Line 29: | ||
===Characterization=== | ===Characterization=== | ||
| − | + | ||
| − | < | + | |
| − | + | <html><p align="justify"> | |
| + | The constructs were cloned into a variant of the pBbB6c<sup>2</sup> medium copy number backbone which lacked the lac-promoter, with the restriction sites <i>EcoRI</i> and <i>XhoI</i>. This backbone has the antibiotic resistance chloramphenicol and the origin of replication (ori) pBBR1. The <i>E. coli</i> strain Top10F was transformed with this biosensor plasmid, as seen in Fig. 3. | ||
</html> | </html> | ||
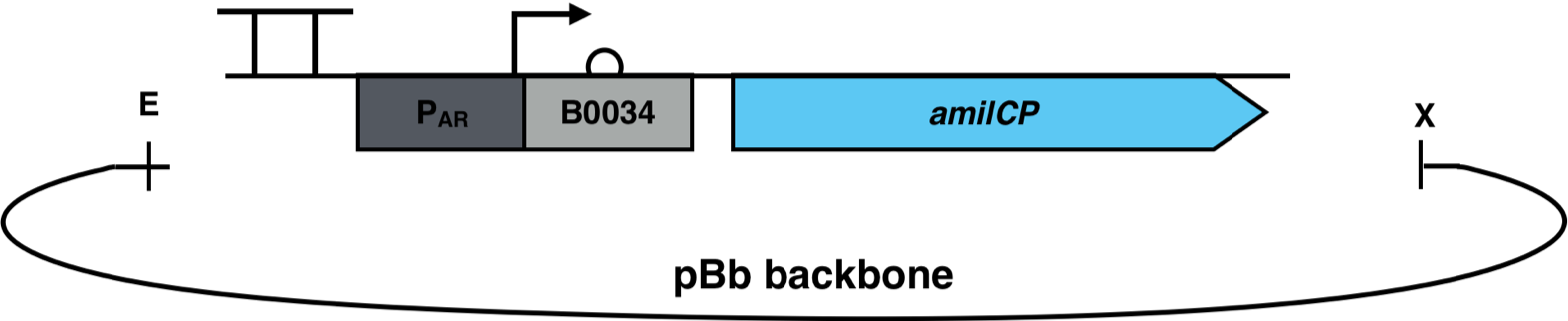
| − | [[File: | + | [[File:AmilCP_Biosensor_scheme.png|500px|thumb|right|<i><b>Fig. 3</b>: P<sub>AR</sub><sup>3</sup> is fused to the reporter gene amilCP and cloned into a pBb backbone. The restriction enzymes EcoRI and XbaI were used for the cloning. The pBb backbone has a chloramphenicol resistance and a medium copy ori pBBR1.</i>]] |
| − | + | ||
| + | <html> | ||
| + | <p align="justify"> | ||
| + | The <a href="https://www.protocols.io/view/transformation-hcnb2ve">transformation</a> of the organism was validated by <a href="https://www.protocols.io/view/e-coli-and-b-subtilis-colony-pcr-8fhhtj6">colony PCR</a>. In some cases, successful transformation could quickly be detected by the different colors of the colonies on the plate and as a cell pellet.<br> | ||
| + | <p align="justify"> | ||
| + | The positive clones were used for experiments and were grown in LB medium over night. The fatty acid stocks for the experiments were prepared. All of the fatty acids were dissolved in ethanol with the exception of butyric acid, which was dissolved in water. The stock solutions of the fatty acids are shown in Table 1. | ||
| + | |||
| + | </html> | ||
| + | <br> | ||
| + | Tab.1: Stock solutions for different fatty acids | ||
| + | <html> | ||
| + | <style> | ||
| + | th { | ||
| + | text-align: left; | ||
| + | } | ||
| + | table, th, td { | ||
| + | border: 1px solid black; | ||
| + | } | ||
| + | </style> | ||
| + | <table style="width:70%"> | ||
| + | <tr> | ||
| + | <th>Fatty acid</th> | ||
| + | <th>Chain length</th> | ||
| + | <th>MW [g/mol]</th> | ||
| + | <th>Solvent</th> | ||
| + | <th>Solubility</th> | ||
| + | <th>Stock concentration</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Butyric acid</td> | ||
| + | <td>C4:0</td> | ||
| + | <td>88.11</td> | ||
| + | <td>Water</td> | ||
| + | <td>60 g/L</td> | ||
| + | <td>200 mM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Capric acid</td> | ||
| + | <td>C10:0</td> | ||
| + | <td>172.26</td> | ||
| + | <td>Ethanol</td> | ||
| + | <td>30 g/L</td> | ||
| + | <td>100 mM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Lauric acid</td> | ||
| + | <td>C12:0</td> | ||
| + | <td>200.32</td> | ||
| + | <td>Ethanol</td> | ||
| + | <td>20 g/L</td> | ||
| + | <td>100 mM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Myristic acid</td> | ||
| + | <td>C14:0</td> | ||
| + | <td>228.37</td> | ||
| + | <td>Ethanol</td> | ||
| + | <td>15 g/L</td> | ||
| + | <td>75 mM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Palmitic acid</td> | ||
| + | <td>C16:0</td> | ||
| + | <td>256.42</td> | ||
| + | <td>Ethanol</td> | ||
| + | <td>20 g/L</td> | ||
| + | <td>75 mM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Stearic acid</td> | ||
| + | <td>C18:0</td> | ||
| + | <td>284.48</td> | ||
| + | <td>Ethanol</td> | ||
| + | <td>20 g/L</td> | ||
| + | <td>50 mM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Oleic acid</td> | ||
| + | <td>C18:1</td> | ||
| + | <td>282.46</td> | ||
| + | <td>Ethanol</td> | ||
| + | <td>100 g/L</td> | ||
| + | <td>200 mM</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </html> | ||
| + | <html><p align="justify"> | ||
| + | The absorption of the overnight cultures was measured and the cultures were used to inoculate fresh LB medium to an OD<sub>600</sub> of 0.05. The cultures were induced with different concentrations of fatty acids. First preliminary tests were carried out with palmitic acid at final concentrations of 0.4 mM, 1 mM and a control without fatty acid. For a better dissolving of the hydrophobic fatty acids in LB medium, Tergitol which was 0.5 % of the whole volume, was added to the medium. The induced samples were transferred on a 24 well plate and the plate was incubated overnight in a 37 °C incubator, which shaked the cultures with 250 rpm. After nearly 16 hours, the samples were taken out of the incubator and distributed on a 96 well plate in 200 µL aliquots. To account for biological heterogeneity and technical errors three biological replicates were measured in three technical replicates each. An empty vector control (EVC) was also supplemented with fatty acids at different concentrations and measured. | ||
| + | All experimental details are listed in Table 2. | ||
| + | <br> | ||
| + | Tab. 2: List of all part improvement experiments | ||
| + | <style> | ||
| + | th { | ||
| + | text-align: left; | ||
| + | } | ||
| + | table, th, td { | ||
| + | border: 1px solid black; | ||
| + | } | ||
| + | </style> | ||
| + | <table style="width:50%"> | ||
| + | <tr> | ||
| + | <th>Construct</th> | ||
| + | <th>Fatty acid</th> | ||
| + | <th>Concentrations</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>P<sub>AR</sub>:amilCP</td> | ||
| + | <td>Lauric acid</td> | ||
| + | <td>0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>P<sub>AR</sub>:amilCP</td> | ||
| + | <td>Myristic acid</td> | ||
| + | <td>0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM</td> | ||
| + | </tr> <tr> | ||
| + | <td>P<sub>AR</sub>:amilCP</td> | ||
| + | <td>Palmitic acid</td> | ||
| + | <td>0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM</td> | ||
| + | </tr> <tr> | ||
| + | <td>P<sub>AR</sub>:amilCP</td> | ||
| + | <td>Stearic acid</td> | ||
| + | <td>0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>EVC</td> | ||
| + | <td>Lauric acid</td> | ||
| + | <td>0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>EVC</td> | ||
| + | <td>Myristic acid</td> | ||
| + | <td>0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>EVC</td> | ||
| + | <td>Palmitic acid</td> | ||
| + | <td>0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>EVC</td> | ||
| + | <td>Stearic acid</td> | ||
| + | <td>0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </html> | ||
| + | |||
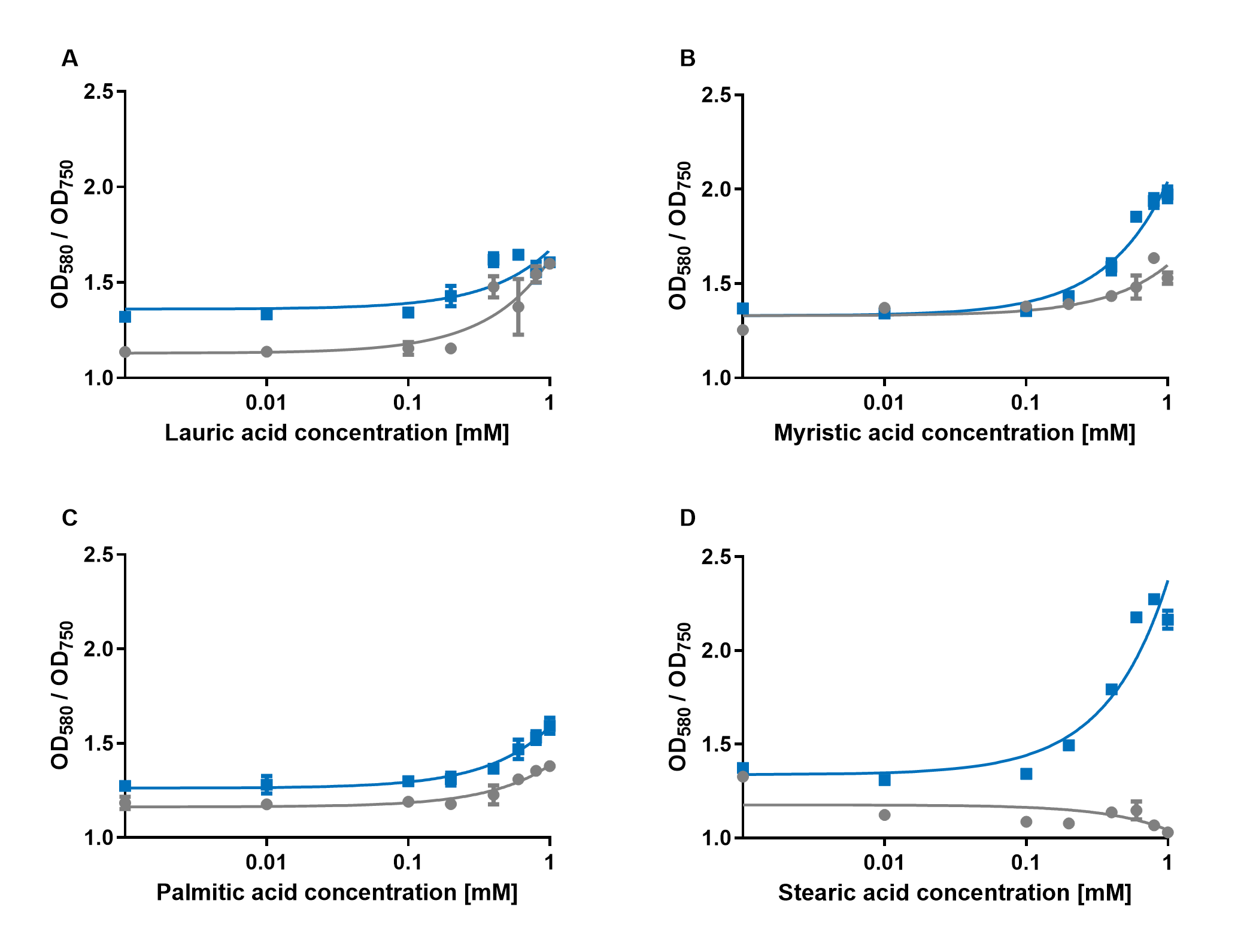
| + | [[File:PAR_AmilCP_ABCD.png|500px|thumb|right|<i><b>Fig.4:</b> Response of P<sub>AR</sub>+amilCP (blue) to different chain lengths of fatty acids compared to an empty vector control (gray). Plot A presents the response for lauric acid, plot B for myristic acid, plot C for palmitic acid and plot D for stearic acid.</i>]] | ||
<html> <p align="justify"> | <html> <p align="justify"> | ||
| − | The experiments with the blue chromoprotein amilCP showed that the production of the chromoprotein is increased by a higher concentration of fatty acids in the medium. The best result was achieved with the fatty acid stearic acid. The biosensor also worked for the chain lengths from C14:0 and C16:0. | + | The experiments with the blue chromoprotein amilCP (<a href="https://parts.igem.org/Part:BBa_K592009">BBa_K592009</a>)<sup>1</sup> showed that the production of the chromoprotein is increased by a higher concentration of fatty acids in the medium. The best result was achieved with the fatty acid stearic acid. The biosensor also worked for the chain lengths from C14:0 and C16:0. Here, the EVC is lower than the samples with amilCP. By adding lauric acid (C12:0) to the culture medium, the production of amilCP increased, but the EVC also increased in a similar manner close to amilCP, so it is not certain if this result can be used for distinct conclusion. |
| + | <p align="justify"> | ||
| + | To summarise the experiments, the created biosensors works for the fatty acids myristic acid (C14:0), palmitic acid (C16:0) and stearic acid (C18:0) with every reporter gene. When using lauric acid (C12:0), the results become more inaccurate. A dose response can still be observed, but it is not as strong or linear as with the other fatty acids. | ||
| + | |||
</html> | </html> | ||
<br><br><br><br><br><br><br><br><br><br> | <br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
===References=== | ===References=== | ||
| − | 1: Fuzhong Zhang, James M Carothers, Jay D Keasling. “Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids” Nature Biotechnology volume 30, pages 354–359 (2012) | + | 1: https://parts.igem.org/Part:BBa_K592009 |
| + | |||
| + | 2: Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. ”BglBrick vectors and datasheets: A synthetic biology platform for gene expression.” J Biol Eng. 2011 Sep 20;5:12. 10.1186/1754-1611-5-12 PubMed 21933410 | ||
| + | |||
| + | 3: Fuzhong Zhang, James M Carothers, Jay D Keasling. “Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids” Nature Biotechnology volume 30, pages 354–359 (2012) | ||
Latest revision as of 19:02, 21 October 2019
Promoter AR with the chromoprotein amilCP
Long-chain fatty acid sensitive promoter PAR expressing amilCP. This part was used to characterize amilCP.
Usage and Biology
The blue chromoprotein amilCP from Acropora millepora (BBa_K592009)1 was used as a reporter gene for a biosensor. The plan was to implement a chromoprotein as a reporter gene, thus enabling to every laboratory to measure a biosensor without fluorescence applications - in contrast, most laboratories have access to a photometer. The chromoprotein amilCP can be observed with the naked eye, in addition to the measurement method by absorption at the wavelength 588 nm for example with a plate reader. Therefore, this option is available for everyone.
The production of long-chain fatty acids (LCFA) is regulated by the transcription factor FadR. This transcription factor can bind to Acyl-CoA which leads to the release of FadR from its cognate operator sequence, thereby increasing gene expression as shown in Fig. 2.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
The constructs were cloned into a variant of the pBbB6c2 medium copy number backbone which lacked the lac-promoter, with the restriction sites EcoRI and XhoI. This backbone has the antibiotic resistance chloramphenicol and the origin of replication (ori) pBBR1. The E. coli strain Top10F was transformed with this biosensor plasmid, as seen in Fig. 3.
The transformation of the organism was validated by colony PCR. In some cases, successful transformation could quickly be detected by the different colors of the colonies on the plate and as a cell pellet.
The positive clones were used for experiments and were grown in LB medium over night. The fatty acid stocks for the experiments were prepared. All of the fatty acids were dissolved in ethanol with the exception of butyric acid, which was dissolved in water. The stock solutions of the fatty acids are shown in Table 1.
Tab.1: Stock solutions for different fatty acids
| Fatty acid | Chain length | MW [g/mol] | Solvent | Solubility | Stock concentration |
|---|---|---|---|---|---|
| Butyric acid | C4:0 | 88.11 | Water | 60 g/L | 200 mM |
| Capric acid | C10:0 | 172.26 | Ethanol | 30 g/L | 100 mM |
| Lauric acid | C12:0 | 200.32 | Ethanol | 20 g/L | 100 mM |
| Myristic acid | C14:0 | 228.37 | Ethanol | 15 g/L | 75 mM |
| Palmitic acid | C16:0 | 256.42 | Ethanol | 20 g/L | 75 mM |
| Stearic acid | C18:0 | 284.48 | Ethanol | 20 g/L | 50 mM |
| Oleic acid | C18:1 | 282.46 | Ethanol | 100 g/L | 200 mM |
The absorption of the overnight cultures was measured and the cultures were used to inoculate fresh LB medium to an OD600 of 0.05. The cultures were induced with different concentrations of fatty acids. First preliminary tests were carried out with palmitic acid at final concentrations of 0.4 mM, 1 mM and a control without fatty acid. For a better dissolving of the hydrophobic fatty acids in LB medium, Tergitol which was 0.5 % of the whole volume, was added to the medium. The induced samples were transferred on a 24 well plate and the plate was incubated overnight in a 37 °C incubator, which shaked the cultures with 250 rpm. After nearly 16 hours, the samples were taken out of the incubator and distributed on a 96 well plate in 200 µL aliquots. To account for biological heterogeneity and technical errors three biological replicates were measured in three technical replicates each. An empty vector control (EVC) was also supplemented with fatty acids at different concentrations and measured.
All experimental details are listed in Table 2.
Tab. 2: List of all part improvement experiments
| Construct | Fatty acid | Concentrations |
|---|---|---|
| PAR:amilCP | Lauric acid | 0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM |
| PAR:amilCP | Myristic acid | 0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM |
| PAR:amilCP | Palmitic acid | 0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM |
| PAR:amilCP | Stearic acid | 0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM |
| EVC | Lauric acid | 0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM |
| EVC | Myristic acid | 0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM |
| EVC | Palmitic acid | 0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM |
| EVC | Stearic acid | 0 mM; 0.01 mM; 0.1 mM; 0.2 mM; 0.4 mM; 0.6 mM; 0.8 mM; 1 mM |
The experiments with the blue chromoprotein amilCP (BBa_K592009)1 showed that the production of the chromoprotein is increased by a higher concentration of fatty acids in the medium. The best result was achieved with the fatty acid stearic acid. The biosensor also worked for the chain lengths from C14:0 and C16:0. Here, the EVC is lower than the samples with amilCP. By adding lauric acid (C12:0) to the culture medium, the production of amilCP increased, but the EVC also increased in a similar manner close to amilCP, so it is not certain if this result can be used for distinct conclusion.
To summarise the experiments, the created biosensors works for the fatty acids myristic acid (C14:0), palmitic acid (C16:0) and stearic acid (C18:0) with every reporter gene. When using lauric acid (C12:0), the results become more inaccurate. A dose response can still be observed, but it is not as strong or linear as with the other fatty acids.
References
1: https://parts.igem.org/Part:BBa_K592009
2: Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. ”BglBrick vectors and datasheets: A synthetic biology platform for gene expression.” J Biol Eng. 2011 Sep 20;5:12. 10.1186/1754-1611-5-12 PubMed 21933410
3: Fuzhong Zhang, James M Carothers, Jay D Keasling. “Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids” Nature Biotechnology volume 30, pages 354–359 (2012)