Difference between revisions of "Part:BBa K861173"
| (7 intermediate revisions by one other user not shown) | |||
| Line 12: | Line 12: | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
__TOC__ | __TOC__ | ||
| Line 21: | Line 21: | ||
=Vilnius-Lithuania 2019 iGEM team contribution= | =Vilnius-Lithuania 2019 iGEM team contribution= | ||
| − | Vilnius-Lithuania 2019 iGEM team | + | Vilnius-Lithuania 2019 iGEM team contributed to the description of Part:BBa_K861173 by measuring growth kinetics of bacteria containing this construct in different conditions. Measuring systems dynamics has superior precision compared to single-point measurements and leads to more straightforward application of data to a mathematical model. In the case of this part characterization, knowing the change in output by varying the glucose concentration, allows the part to be used in controlling the protein of interest level in the cell. |
| + | |||
| + | <b>Method</b> | ||
| + | |||
| + | For measurements, the iGEM community protocol suggests using LB media. From our experience, a significant drawback is its considerable autofluorescence, which restricts measurement precision, especially for low expression systems. Also, performing measurements in M9 media allows more control in varying the growth environment. For these reasons, we chose to use M9-CA media. | ||
| + | |||
| + | We measured growth kinetics with varying glucose (5%, 2%, 1% 0.4%, 0.1%, 0.01%, 0.001%) and glycerol (0.4%) concentrations by mass. The measurements were done in a microplate reader, points were taken every 30 minutes for 14 hours. | ||
| + | |||
| + | Measurements were conducted at 37°C, E. coli DH5α strain, measurement volume – 100 μL. Two measurements for two different colonies were done. | ||
| + | |||
| + | R code was written for data analysis and visualization. | ||
| + | |||
| + | Full experimental protocol and microplate reader settings can be found [https://2019.igem.org/wiki/images/2/24/T--Vilnius-Lithuania--Measurement-PDF.pdf here.] | ||
| + | |||
| + | Full R code can be found [https://2019.igem.org/wiki/images/c/c0/T--Vilnius-Lithuania--Measurement-R.pdf here.] | ||
| + | |||
| + | <b>The results</b> | ||
| + | |||
| + | A 9 fold difference was observed between bacteria, grown in M9-CA, containing 5% and 0% glucose. Also, a difference in output, when grown in glucose and glycerol containing media can be seen, which can be explained by the difference in by the metabolic pathways used by the tested cells. | ||
| + | [[Image:T--Vilnius-Lithuania--K861173.png|thumb|centre|900px|<b>Figure 1.</b>The graph showing change in mRFP expression in time with different glucose concentrations (5%, 2%, 1% 0.4%, 0.1%, 0.01%, 0.001%) and glycerol (0.4%). A 9 fold difference can be seen between bacetria grown in 5% and 0% glucose media. The measuring was conducted in 37°C, M9-CA media, E. coli DH5α strain.]] | ||
| − | + | Vilnius-Lithuania 2019 iGEM team suggests future teams that choose this part for characterization to measure kinetics with different carbon sources and varying their concentrations. | |
='''IISER Kolkata 2019'''= | ='''IISER Kolkata 2019'''= | ||
| Line 107: | Line 126: | ||
concentration of glucose, fructose and xylose. | concentration of glucose, fructose and xylose. | ||
</p> | </p> | ||
| + | |||
| + | |||
| + | |||
| + | ==Functional Parameters: Austin_UTexas== | ||
| + | <html> | ||
| + | <body> | ||
| + | <partinfo>BBa_K861173 parameters</partinfo> | ||
| + | <h3><center>Burden Imposed by this Part:</center></h3> | ||
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/f/fa/T--Austin_Utexas--no_burden_icon.png" style = "width:160px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: 3.1 ± 2.6% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </body> | ||
| + | </html> | ||
Latest revision as of 19:11, 3 September 2020
mRFP generator controlled by PcstA (glucose-repressible promoter)
RFP is controlled by promoter PcstA, which can be activated by CRP-cAMP complex. Therefore RFP will be expressed at low glucose concentration.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 712
Illegal AgeI site found at 824 - 1000COMPATIBLE WITH RFC[1000]
Contents
Vilnius-Lithuania 2019 iGEM team contribution
Vilnius-Lithuania 2019 iGEM team contributed to the description of Part:BBa_K861173 by measuring growth kinetics of bacteria containing this construct in different conditions. Measuring systems dynamics has superior precision compared to single-point measurements and leads to more straightforward application of data to a mathematical model. In the case of this part characterization, knowing the change in output by varying the glucose concentration, allows the part to be used in controlling the protein of interest level in the cell.
Method
For measurements, the iGEM community protocol suggests using LB media. From our experience, a significant drawback is its considerable autofluorescence, which restricts measurement precision, especially for low expression systems. Also, performing measurements in M9 media allows more control in varying the growth environment. For these reasons, we chose to use M9-CA media.
We measured growth kinetics with varying glucose (5%, 2%, 1% 0.4%, 0.1%, 0.01%, 0.001%) and glycerol (0.4%) concentrations by mass. The measurements were done in a microplate reader, points were taken every 30 minutes for 14 hours.
Measurements were conducted at 37°C, E. coli DH5α strain, measurement volume – 100 μL. Two measurements for two different colonies were done.
R code was written for data analysis and visualization.
Full experimental protocol and microplate reader settings can be found here.
Full R code can be found here.
The results
A 9 fold difference was observed between bacteria, grown in M9-CA, containing 5% and 0% glucose. Also, a difference in output, when grown in glucose and glycerol containing media can be seen, which can be explained by the difference in by the metabolic pathways used by the tested cells.

Vilnius-Lithuania 2019 iGEM team suggests future teams that choose this part for characterization to measure kinetics with different carbon sources and varying their concentrations.
IISER Kolkata 2019
The Characterization of BBa_K381001
BBa_K861173 has a mRFP controlled by PcstA which is a Glucose repressible promoter. It has been shown by previous teams that with the increased concentration of glucose there is a decrease in the activity of PcstA. Team iGEM IISER Kolkata is trying to characterize what is the effect of increasing concentration of other glucose analogues on the same system.
Aim
BBa_K861173 was used to characterize the promoter activity of PcstA in a range of concentration of glucose and its analogues in the growth media.
Method
E. coli DH5α were transformed using re-suspended BBa_K861173 pSB1C3 plasmids from iGEM 2019 distribution kit and selected using Chloramphenicol LB agar plates.
- Single positive colony of BBa_I13522 was added to 10ml of LB media and left overnight at 37° C on 150 rpm in an incubator. Along with this, a positive control with non-transformed E coli cells was also kept in liquid culture in the similar conditions.
- 200 µL of overnight culture was added to 10ml of M9 minimal media with different concentrations of sugars in individual test tubes. One control was kept in LB media.
- It was kept in incubated for 8 hours at 37°C at 150 rpm.
- At the end of 8 hours of incubation in the incubator, 100uL of the culture was added to each well of 96 well plate. The culture in LB was pelleted down and resuspended in equivalent amount of M9 media.
- Varioskan LUX multimode reader was used to measure the GFP fluorescence: Excitation at 494 nm and emission at 525 nm.
- Additionally, Shimazadu UV1900 was used to measure the OD at 600 nm to quantify the bacterial growth.
Result
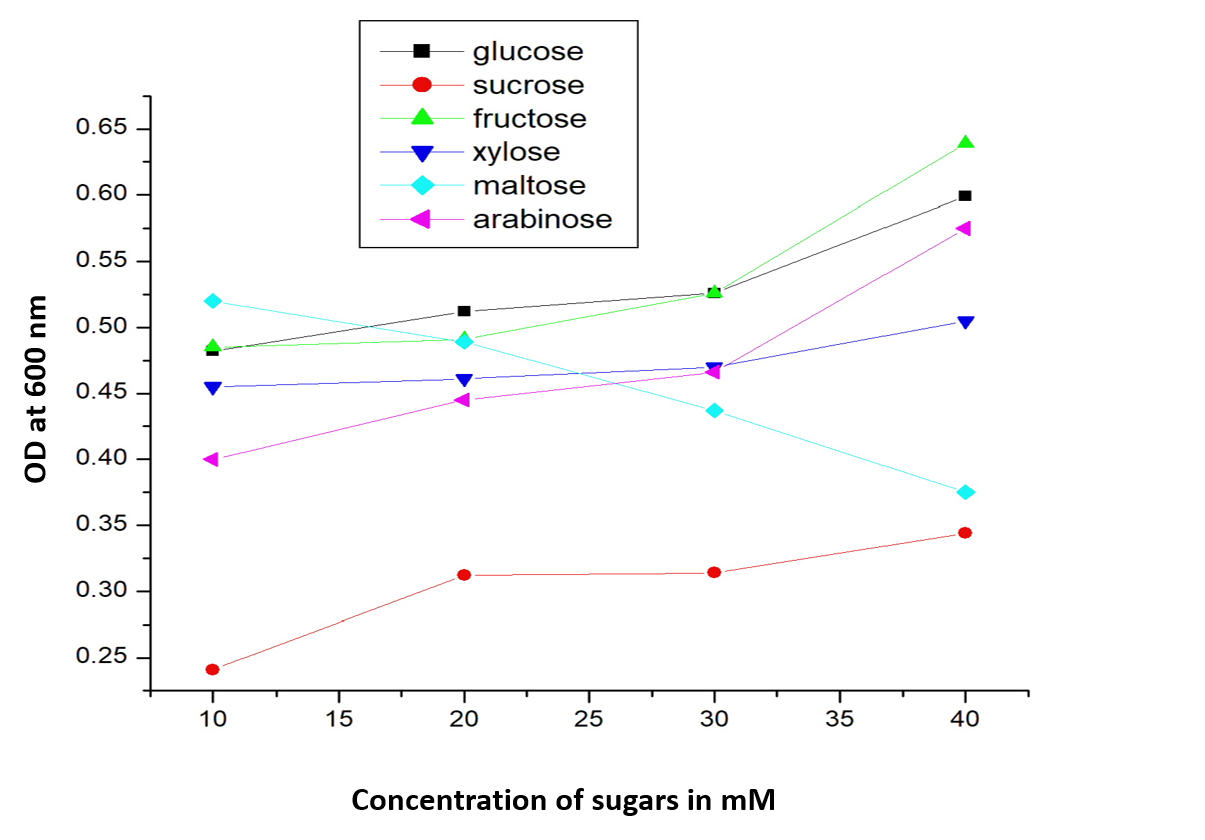 OD at 600 nm for E coli transformed with BBa_I13522 grown in range of concentrations (10 mM, 20mM, 30mM and 40mM) of glucose and its analogues
OD at 600 nm for E coli transformed with BBa_I13522 grown in range of concentrations (10 mM, 20mM, 30mM and 40mM) of glucose and its analogues

Decrease in mRFP fluorescence with increase in concentration of glucose, fructose and xylose. The data for rest three sugars viz. sucrose, maltose and arabinose has not been shown as they show no clear co-relation of mRFP production and sugar concentration
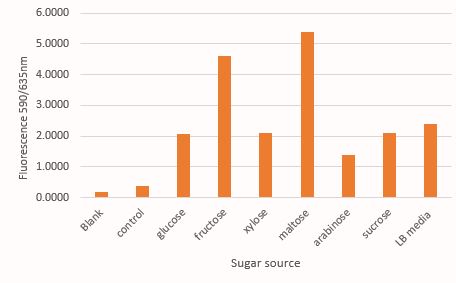
Comparison of mRFP fluorescence in M9 media supplemented with different sugar source at 30mM concentration. Here the control contains untransformed DH5α grown in LB media under a similar condition like that of test samples
Conclusion
As expected, there is a linear increase in the OD600 of the bacterial culture with the increase in the concentration of sugar (except maltose) as seen in figure 1. Moreover, we could also show a clear co-relation that with the increase in concentration of glucose, fructose and xylose there is a decrease in mRFP fluorescence. This signifies the fact that the PcstA promoter activity decreases with the increase in concentration of glucose, fructose and xylose.
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
