Difference between revisions of "Part:BBa K516132"
| (6 intermediate revisions by 3 users not shown) | |||
| Line 9: | Line 9: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | |||
| + | ===Characterization=== | ||
| + | |||
| + | ==iGEM_BIT 2019’s Characterisation== | ||
| + | In 2019, IGEM_BIT characterized the BBa-K516132 in the microfluidic chip . | ||
| + | |||
| + | More details can be found at experience page :https://parts.igem.org/Part:BBa_K516132:Experience | ||
| + | |||
==Shanghai_High_School 2019’s Characterisation== | ==Shanghai_High_School 2019’s Characterisation== | ||
==BBa_K516132 Constitutive promoter [J23101] with mRFP, RBS B0032== | ==BBa_K516132 Constitutive promoter [J23101] with mRFP, RBS B0032== | ||
| Line 24: | Line 32: | ||
<p>We measured the RFU/OD600 of the strain at 24h and 30h, respectively. The RFU/OD600 at 24h is 49.57 ,which is higher than the RFU/OD600 31.04 at 30h. </p> | <p>We measured the RFU/OD600 of the strain at 24h and 30h, respectively. The RFU/OD600 at 24h is 49.57 ,which is higher than the RFU/OD600 31.04 at 30h. </p> | ||
| − | [[Image:K516132- | + | [[Image:K516132-3.png|400px|Characterization of popular BioBrick RBSs]] |
<strong>Figure3. BBa_K516132 containing strain Fluorescence per OD600 at 30℃ and 37℃. </strong> | <strong>Figure3. BBa_K516132 containing strain Fluorescence per OD600 at 30℃ and 37℃. </strong> | ||
| Line 40: | Line 48: | ||
| − | |||
===Functional Parameters=== | ===Functional Parameters=== | ||
<partinfo>BBa_K516132 parameters</partinfo> | <partinfo>BBa_K516132 parameters</partinfo> | ||
| − | < | + | |
| + | |||
| + | |||
| + | ==Functional Parameters: Austin_UTexas== | ||
| + | <html> | ||
| + | <body> | ||
| + | <h3><center>Burden Imposed by this Part:</center></h3> | ||
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/4/43/T--Austin_Utexas--Low_significant_burden.png" style = "width:200px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: 8.7 ± 4.8% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This part exhibited a significant burden. Users should be aware that BioBricks with a burden of >20-30% may be susceptible to mutating to become less functional or nonfunctional as an evolutionary consequence of this fitness cost. This risk increases as they used for more bacterial cell divisions or in larger cultures. Users should be especially careful when combining multiple burdensome parts, as plasmids with a total burden of >40% are expected to mutate so quickly that they become unclonable. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods and other conclusions from a large-scale measurement project conducted by the <a href="https://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team.</a></p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </p> | ||
| + | </body> | ||
| + | </html> | ||
Latest revision as of 00:26, 4 September 2020
Constitutive promoter [J23101] with mRFP, RBS B0032
Protein Generator regulated by BBa_J23101 constitutive promoter with BBa_B0032 medium efficiency RBS, mRFP Reporter Coding Region and a double Transcription Terminators.
BBa_J23101 is the reference standard promoter for the computation of RPUs.
This measurement device belongs to a set of parts useful to estimate the RBS efficiency (BBa_K516130; BBa_K516131).
Usage and Biology
Characterization
iGEM_BIT 2019’s Characterisation
In 2019, IGEM_BIT characterized the BBa-K516132 in the microfluidic chip .
More details can be found at experience page :https://parts.igem.org/Part:BBa_K516132:Experience
Shanghai_High_School 2019’s Characterisation
BBa_K516132 Constitutive promoter [J23101] with mRFP, RBS B0032
The change in fluorescence intensity in the medium is related to the culture conditions, such as culture time, culture temperature, etc., in addition to the type of fluorescent protein. We studied this by experimenting with changes in culture time, culture temperature and fluorescence protein type.
1. We transformed the plasmid contanining BBa_K516132 and plasmid contanining BBa_K1073024 into bacterial competent cells separately, plated and cultured overnight at 37 °C.
2. On the evening of the next day, we picked single clone into a tube containing 5 ml LB which is called 0h. At the same time, we also inoculated competent DH5α strain into a tube containing 5 ml LB as a blank control. We culture bacterial cells at 37 ° C, 220 rpm. The sampling time point is 24h.
3. We took 100ul of the bacteria culture and measured the total fluorescence with a Thermo fluorescence microplate reader. We took 200ul of the ten-fold diluted bacterial solution and measured the OD600 with BioTek optical microplate reader. Total fluorescence is divided by OD600 to obtain fluorescence value per OD600.
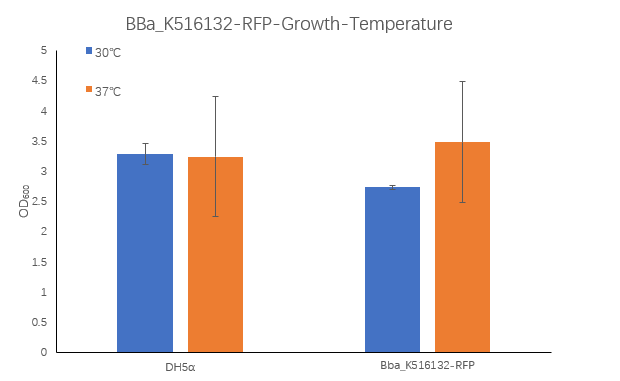
Figure1. BBa_K516132 containing strain OD600 at 30℃ and 37℃.
The OD600 of blank control DH5α cells grown at 30 ° C and 37 ° C were 3.285 and 3.245, respectively, which is very close. The OD600 of the strain containing BBa_K516132 grown at 30 ° C was 2.735, which was lower than the OD600 3.485 at 37 ° C.
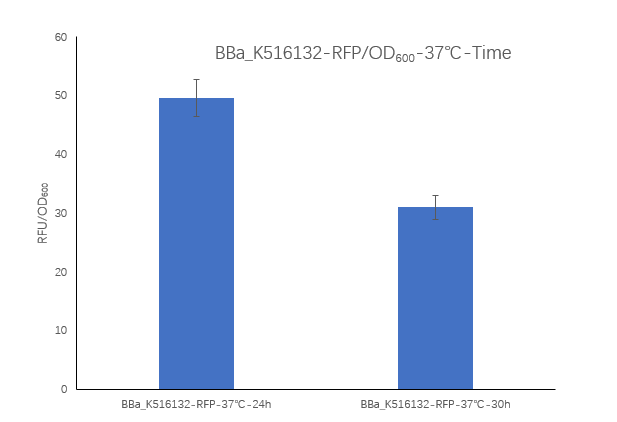
Figure2. Ba_K516132 containing strain Fluorescence per OD600 at 24h and 30h.
We measured the RFU/OD600 of the strain at 24h and 30h, respectively. The RFU/OD600 at 24h is 49.57 ,which is higher than the RFU/OD600 31.04 at 30h.
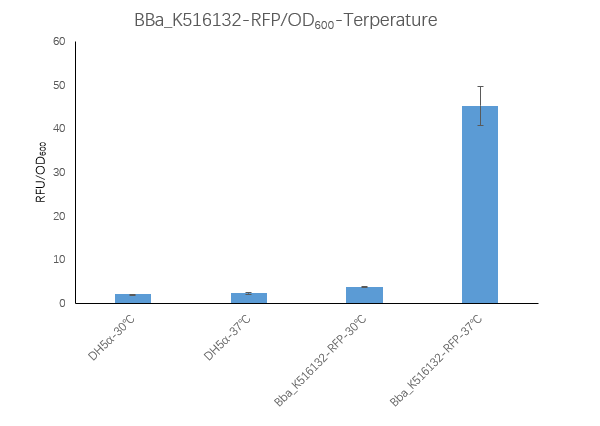
Figure3. BBa_K516132 containing strain Fluorescence per OD600 at 30℃ and 37℃.
We measured the RFU/OD600 of the strain at 30℃and 37℃, respectively. The DH5α RFU/OD600 at 24h and 30h is close to zero. The Bba_K516132 containing strain
RFU/OD600 is 45.26 at 37℃,which is higher than 3.74 at 30℃.
To sum up: the growth OD600 of the BBa_K516132-RFP strain is similar to the control DH5α, and this strain should have reached a stable phase at 24 h. The BBa_K516132-RFP strain has a much higher fluorescence at 37 ° C than 30 ° C.
These results indicate that the BBa_K516132-RFP protein may be expressed more or more stably at 37 °C, which also means that it may be necessary to find a suitable culture temperature when expressing an unknown protein.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 617
Illegal AgeI site found at 729 - 1000COMPATIBLE WITH RFC[1000]
Functional Parameters
| n/a | Constitutive promoter [J23101] with mRFP, RBS B0032 |
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This part exhibited a significant burden. Users should be aware that BioBricks with a burden of >20-30% may be susceptible to mutating to become less functional or nonfunctional as an evolutionary consequence of this fitness cost. This risk increases as they used for more bacterial cell divisions or in larger cultures. Users should be especially careful when combining multiple burdensome parts, as plasmids with a total burden of >40% are expected to mutate so quickly that they become unclonable. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.