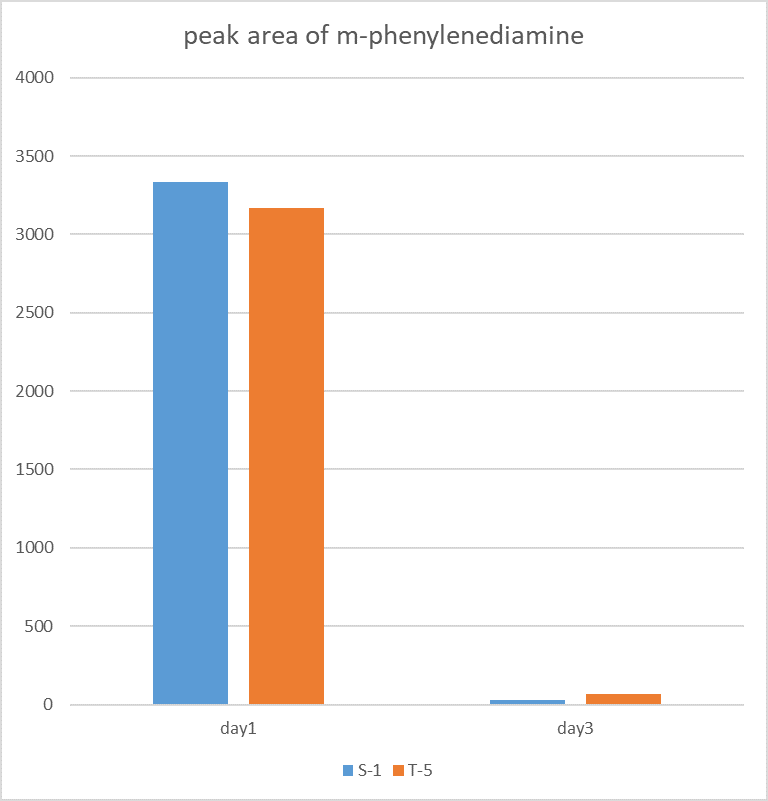Difference between revisions of "Part:BBa K863001"
Duduu36lllf (Talk | contribs) |
|||
| (2 intermediate revisions by one other user not shown) | |||
| Line 4: | Line 4: | ||
bpul (Laccase from Bacillus pumilus) | bpul (Laccase from Bacillus pumilus) | ||
| − | + | ===Usage and Biology=== | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
First some trials of shaking flask cultivations were made with different parameters to define the best conditions for production of the His tagged CotA aus [http://www.dsmz.de/catalogues/details/culture/DSM-27.html ''Bacillus pumilus'' DSM 27 ( ATCC7061)] named BPUL. Due to inactivity of the enzyme in the cell lysate a purification method was established (using Ni-NTA-Histag resin and Syringe or ÄKTA method). The purified BPUL could be detected by SDS-PAGE (molecular weight of 58.6 kDa) as well as by MALDI-TOF. To improve the purification strategies the length of the linear elution gradient was increased up to 200 mL . The fractionated samples were also tested concerning their activity and revealed high activity. Optimal conditions for activity were identified. After measuring activity of BPUL a successful scale up was made up to 3 L and also up to 6 L that enables an intense screening afterwards. The scale up will be important for a further application. | First some trials of shaking flask cultivations were made with different parameters to define the best conditions for production of the His tagged CotA aus [http://www.dsmz.de/catalogues/details/culture/DSM-27.html ''Bacillus pumilus'' DSM 27 ( ATCC7061)] named BPUL. Due to inactivity of the enzyme in the cell lysate a purification method was established (using Ni-NTA-Histag resin and Syringe or ÄKTA method). The purified BPUL could be detected by SDS-PAGE (molecular weight of 58.6 kDa) as well as by MALDI-TOF. To improve the purification strategies the length of the linear elution gradient was increased up to 200 mL . The fractionated samples were also tested concerning their activity and revealed high activity. Optimal conditions for activity were identified. After measuring activity of BPUL a successful scale up was made up to 3 L and also up to 6 L that enables an intense screening afterwards. The scale up will be important for a further application. | ||
| Line 251: | Line 247: | ||
| + | ==Xiamen_City 2019's characterization== | ||
| + | ==BBa_K863001 bpul laccase from Bacillus pumilus== | ||
| + | ===(1) Transformation of Plasmid DNA=== | ||
| + | <strong>Material: Plasmid (pSB1C3-J23100-B0034-bpul-ter)</strong> | ||
| + | <p>1. Take out E. coli competent cell (MG1655, DH10B, BL21(DE3)) from the refrigerator</p> | ||
| + | <p>2. Put the plasmid on ice and put it in a constant temperature water bath (42 Celsius) for 90 s</p> | ||
| + | <p>3. Use the needle tip to take a small amount of plasmid, add it to the tube with 300 μL medium, and put it onto the plate with solid media.</p> | ||
| + | <p>4. Pick the colonies in LB media and put it in shaker at 37℃</p> | ||
| + | |||
| + | ===(2) Protein Electrophoresis=== | ||
| + | <strong>Material: MG1655 bpul, DH10B bpul, BL21(DE3) bpul, loading solution (6 μL), deionized water (1000 μL), marker solution, Coomassie brilliant blue dye.</strong> | ||
| + | <p>1. Add 24 μL MG1655 DH10B bpul, BL21(DE3) bpul to 1.5mL tube respectively, and then add 6 μL loading.</p> | ||
| + | <p>2. Add 500 μL deionized water and centrifuge the tubes respectively.</p> | ||
| + | <p>3. Add another 500 μL deionized water, blow and suspend the solution. </p> | ||
| + | <p>4. Put the tubes 99℃ water for 15 minutes.</p> | ||
| + | <p>5. After heating, centrifuge the tubes for 5min.</p> | ||
| + | <p>6. Take and install the gel, add the electrophoresis buffer to the sample hole, and check if there is any leakage. Add marker, to the first and last sample holes, add MG1655 bpul, DH10B bpul, BL21(DE3) bpul to the other sample holes, respectively.</p> | ||
| + | <p>7. Connect the electrophoresis device to the power supply. Connect the positive electrode to the tank and the negative electrode to the slot for electrophoresis. The voltage is adjusted to 160V. </p> | ||
| + | <p>8. Turn off the power supply and disconnect the electrode until the bromophenol blue reaches the bottom of the release adhesive. Remove the glass sheet from the electrophoresis device and then remove the gel. </p> | ||
| + | <p>9. Soak the gel in Coomassie brilliant blue dye and dye it in a horizontal shaking bed for 15 min.</p> | ||
| + | |||
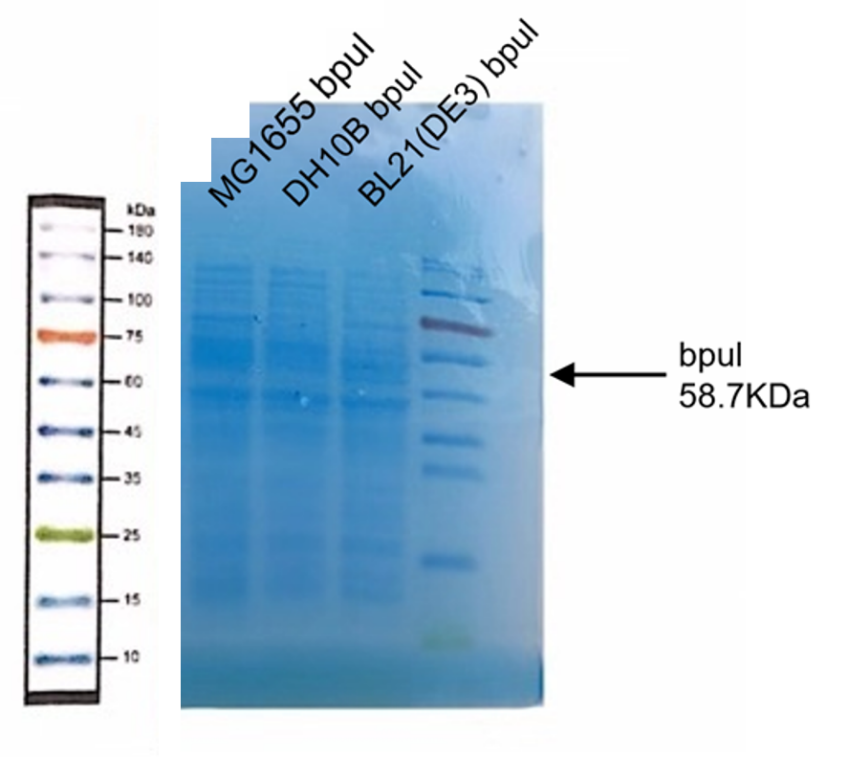
| + | <p>Observe the protein bands</p> | ||
| + | [[Image:K3158005-1.png|400px|Characterization of popular BioBrick RBSs]] | ||
| + | |||
| + | ===(3) HPLC === | ||
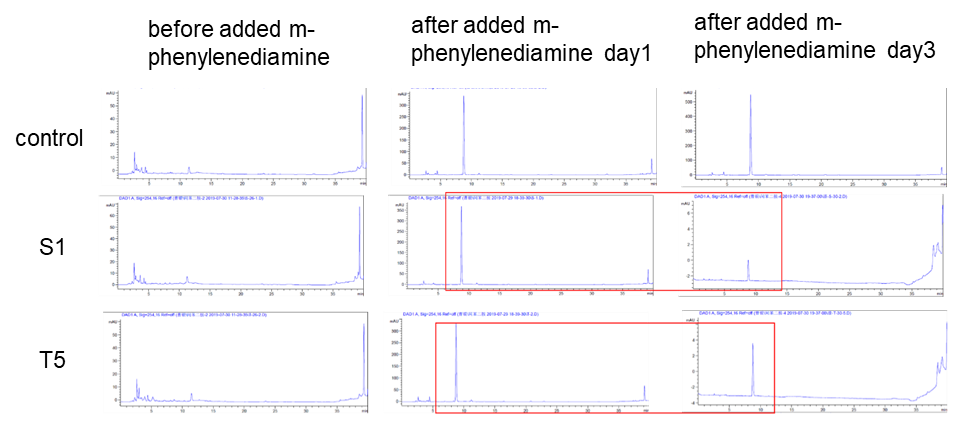
| + | <p>Two colonies (DH10B bpul) were picked and added with 5000 um of m-phenylenediamine, and the left side in the figure was a spectrum with nothing added. The middle was added with m-phenylenediamine for one day, and the right was after adding m-phenylenediamine for three days, the content of S-1 and T-5 was found to decrease when the content was detected.</p> | ||
| + | [[Image:K3158005-2.png|400px|Characterization of popular BioBrick RBSs]] | ||
| + | <p>The figure shows the peak area of m-phenylenediamine that detected for m-phenylenediamine</p> | ||
| + | [[Image:K3158005-3.png|400px|Characterization of popular BioBrick RBSs]] | ||
| + | |||
| + | =Contribution From GEC-Beijing = | ||
| + | ==Description== | ||
| + | We modified the existing part accase gene BBa_K863001 using surface display technology, resulting in a new part INP-Bpul (BBa_K5458001). Considering the unsafe and time-consuming process of jeans bleaching technology currently used in the market, we designed to use laccase to degrade the indigo dye in jeans to achieve more precise control of the degree of jeans bleaching and reduce damage to the fabric. We chose the laccase gene Bpul from Bacillus pumilus. Laccase has many functions, such as decolorizing textiles, removing harmful substances from wastewater, and degrading many industrial pollutants. | ||
| + | |||
| + | ==Usage and Biology== | ||
| + | We first selected and codon-optimized the laccase gene Bpul (BBa_K863001) from Bacillus pumilus to ensure efficient expression in E. coli. To improve expression efficiency, we optimized the gene sequence and sent it to a biotechnology company for synthesis. Next, the synthesized Bpul gene was inserted into the pET23b expression vector via EcoRI and XhoI double digestion. The Bpul gene was precisely positioned at the multiple cloning site (MCS) of the vector, ensuring efficient expression . After constructing the recombinant plasmid, it was transformed into E. coli BL21, and positive clones were confirmed through antibiotic selection. Ultimately, we successfully obtained the engineered strain BL21/pET23b-Bpul, which expresses the laccase. | ||
| + | |||
| + | <html> | ||
| + | <div style="display:flex; flex-direction: column; align-items: center;"> | ||
| + | <img src="https://static.igem.wiki/teams/5458/gene-ciruit-bpul.png" style="width: 500px;margin: 0 auto" /> | ||
| + | <p style="font-size: 98%; line-height: 1.4em;">Figure 1. The gene circuit of Bpul.</p > | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | ==Characterization== | ||
| + | ===Laccase activity assay=== | ||
| + | |||
| + | <html> | ||
| + | <div style="display:flex; flex-direction: column; align-items: center;"> | ||
| + | <img src="https://static.igem.wiki/teams/5458/gel-image-of-the-bgls-and-bupl.png" style="width: 500px;margin: 0 auto" /> | ||
| + | <p style="font-size: 98%; line-height: 1.4em;">Figure 2. Gel image of the Bgls and Bupl. The Bgls gene is related to cellulase and will be studied in later experiments.</p > | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | The experimental procedure was as follows: 100 mL of overnight culture of the engineered strain was collected, and its OD600 was measured. After centrifugation at 10,000 rpm for 1 min, the bacterial cells were resuspended in 20 mL of Britton-Robinson (BR) buffer and then sonicated to disrupt the cells. The supernatant was collected after centrifuging at 10,000 rpm for 20 min at 4℃, yielding the crude enzyme solution. 100 μL of the crude enzyme solution was taken, and the total protein concentration was determined using the Bradford assay, adjusting the concentration to 100 μg/mL. To assess laccase activity, 1 mM ABTS was used as the substrate, and the reaction was incubated at 37℃. The oxidation of ABTS, indicated by an increase in absorbance at 420 nm, was measured using a microplate reader. The results demonstrate that the Bpul gene from Bacillus pumilus was successfully expressed in E. coli BL21 and exhibited strong laccase activity. Compared to the control groups, the engineered strain expressing laccase significantly accelerated the degradation of ABTS, confirming its potential for degrading indigo dye in denim applications. | ||
| + | |||
| + | <html> | ||
| + | <div style="display:flex; flex-direction: column; align-items: center;"> | ||
| + | <img src="https://static.igem.wiki/teams/5458/bpul-abts.png" style="width: 500px;margin: 0 auto" /> | ||
| + | <p style="font-size: 98%; line-height: 1.4em;">Figure 3. Absorbance at 420nm for different strains. As shown in the Figure 3, the absorbance values at 420 nm for the wild-type BL21, the BL21/pET23b empty vector, and the BL21/p23b-Bpul strains were compared. Both the wild-type BL21 and the BL21/pET23b strains showed negligible laccase activity, with absorbance values close to zero. In contrast, the BL21/p23b-Bpul strain exhibited a significant increase in absorbance, reaching 2.17, indicating that the expressed laccase effectively catalyzed the oxidation of ABTS.</p > | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | ===Laccase catalytic time assay=== | ||
| + | First, 100 μL of crude enzyme solution was collected, and the total protein concentration was measured using the Bradford protein assay. The protein concentration was adjusted to 100 μg/mL using BR buffer. Then, 1 mM ABTS was added to 200 μL of the enzyme solution. The reaction mixture was incubated at 37℃ in a sealed environment, and the absorbance at 420 nm was measured at different time points: 5 min, 10 min, 15 min, 20 min, 25 min, and 30 min, using a microplate reader.The optimal catalytic time forlaccase is 15 min. This finding is valuable for applications in denim treatment, as it helps optimize the enzyme’s use in degrading indigo dye efficiently within a controlled timeframe. | ||
| + | |||
| + | <html> | ||
| + | <div style="display:flex; flex-direction: column; align-items: center;"> | ||
| + | <img src="https://static.igem.wiki/teams/5458/degradation-by-laccase-over-time.png" style="width: 500px;margin: 0 auto" /> | ||
| + | <p style="font-size: 98%; line-height: 1.4em;">Figure 4. Absorbance at 420 nm for ABTS degradation by laccase over time. As shown in Figure 4, the absorbance values at 420 nm, reflecting the degradation of ABTS by laccase, varied across the different time intervals. The control group (wild-type E. coli BL21) exhibited absorbance values close to zero throughout the experiment. In contrast, the E. coli BL21-Bpul strain expressing laccase showed a rapid increase in absorbance during the early stages of the reaction. The absorbance reached its peak at approximately 2.05 at 15 min. After 15 min, the absorbance remained stable.</p > | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | ===Influence of temperature on laccase activity=== | ||
| + | 100 μL of previously prepared crude enzyme solution was used. The protein concentration was measured using the Bradford protein assay and adjusted to 100 μg/mL. A 100 mM ABTS solution was prepared with distilled water, and 1 mM ABTS was added to 200 μL of the enzyme solution. Laccase activity was tested at different temperatures (25℃, 37℃, 45℃, 55℃, 70℃) using a thermostat water bath. The absorbance at 420 nm was measured over a 30-min period to assess ABTS degradation.The results showed that the optimal temperature for laccase activity is 55℃, where the enzymatic reaction efficiency is highest. | ||
| + | |||
| + | <html> | ||
| + | <div style="display:flex; flex-direction: column; align-items: center;"> | ||
| + | <img src="https://static.igem.wiki/teams/5458/laccase-activity-at-different-temperatures.png" style="width: 500px;margin: 0 auto" /> | ||
| + | <p style="font-size: 98%; line-height: 1.4em;">Figure 5. Laccase activity at different temperatures. The results showed that laccase activity increased with rising temperature and reached its peak at 55℃, with an absorbance of 2.93. Beyond 55℃, the activity dropped sharply, with absorbance falling to 1.65 at 70℃.</p > | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | ===Influence of pH on laccase activity=== | ||
| + | The engineered bacteria were resuspended in BR buffer with varying pH values from 4 to 9. After disrupting the cells, the supernatant containing the crude enzyme was collected by centrifugation at 10,000 rpm for 20 min at 4℃. Then, 1 mM ABTS was added to the enzyme solution, and the reactions were incubated at 37℃ for 30 min in a sealed environment. Absorbance at 420 nm was measured using a microplate reader to assess laccase activity.The experimental results indicate that the optimal pH for laccase activity is pH=5, where the enzyme exhibits the highest catalytic efficiency. | ||
| + | |||
| + | <html> | ||
| + | <div style="display:flex; flex-direction: column; align-items: center;"> | ||
| + | <img src="https://static.igem.wiki/teams/5458/laccase-activity-at-different-ph.png" style="width: 500px;margin: 0 auto" /> | ||
| + | <p style="font-size: 98%; line-height: 1.4em;">Figure 6. Laccase activity at different pH. As shown in Figure 6, the activity of laccase varied across the pH range tested. The enzyme's activity increased as the pH rose from 4 to 5, reaching its peak at pH=5, with absorbance value of approximately 2.65. Beyond pH=5, the activity decreased rapidly, indicating that the enzyme becomes less efficient as the pH increases toward neutral and alkaline conditions.</p > | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | ===Influence of copper irons on laccase activity=== | ||
| + | This experiment aims to study the effect of copper ions on laccase activity, as copper ions serve as the active center of laccase. By determining the optimal copper ion concentration, we can assess whether adding copper chloride to the final product can enhance the enzyme's catalytic efficiency. A 100 mM CuCl2 solution was prepared, and different concentrations of CuCl2 (0, 0.1, 0.25, 0.5, and 1 mM) were added to the ABTS catalytic reaction system to evaluate the effect of copper ions on laccase activity. The reactions were incubated at 37℃, pH=5, for 30 min, and the absorbance at 420 nm was measured to assess enzyme activity.The results indicate that 0.5 mM is the optimal copper ion concentration to enhance laccase activity. Based on this finding, we can add an appropriate concentration of copper ions to our laccase products to improve enzymatic activity. | ||
| + | |||
| + | <html> | ||
| + | <div style="display:flex; flex-direction: column; align-items: center;"> | ||
| + | <img src="https://static.igem.wiki/teams/5458/different-concentrations-of-copper-ions.png" style="width: 500px;margin: 0 auto" /> | ||
| + | <p style="font-size: 98%; line-height: 1.4em;">Figure 7. Effect of different concentrations of copper ions on laccase activity.The experimental results showed that laccase activity increased as the copper ion concentration increased from 0 mM to 0.5 mM, with the highest activity observed at 0.5 mM. However, when the concentration increased to 1 mM, laccase activity decreased.</p > | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | ===Testing the degradation of indigo by laccase=== | ||
| + | Previous experiments demonstrated laccase activity but did not confirm the enzyme's ability to degrade indigo. Therefore, this experiment aims to test laccase's effectiveness in degrading indigo dye. To improve the solubility of indigo, 10 mM indigo (SM4168, Beyotime) was dissolved in dimethyl sulfoxide (DMSO). A 1 mM indigo solution was added to 200 μL of crude enzyme samples (BR buffer, pH=5). A standard curve was established using different concentrations of indigo (0 mM, 0.25 mM, 0.5 mM, 1 mM, 1.5 mM, and 3 mM). The experiment was performed in a 96-well plate, sealed, and incubated at 37℃ for 2 hours. Each group was done in triplicate, and the optical density was measured at 620 nm, with indigo concentrations calculated based on the standard curve.The experimental results demonstrate that the BL21/Bpul strain, which expresses the laccase gene, significantly degrades indigo more effectively than the BL21 control strain. This confirms that laccase has strong catalytic activity in indigo degradation. | ||
| + | |||
| + | <html> | ||
| + | <div style="display:flex; flex-direction: column; align-items: center;"> | ||
| + | <img src="https://static.igem.wiki/teams/5458/effect-of-laccase-on-indigo-degradation.png" style="width: 500px;margin: 0 auto" /> | ||
| + | <p style="font-size: 98%; line-height: 1.4em;">Figure 8. Effect of laccase on indigo degradation. As shown in Figure 8, after 2 hours of incubation, the indigo concentration in the BL21 strain averaged 0.95 mM, while in the BL21/Bpul strain it averaged 0.51 mM. The indigo concentration in the BL21 strain was significantly higher than that in the BL21/Bpul strain, indicating that the BL21/Bpul strain has a stronger ability to degrade indigo. </p > | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | ==Potential Application Directions== | ||
| + | |||
| + | This experiment demonstrated that the Bpul gene from Bacillus pumilus exhibits the highest laccase activity under the following conditions: temperature of 55°C, pH=5, copper ion concentration of 0.5 mM, and substrate ABTS concentration of 0.5 mM, with an optimal reaction time of 15 minutes. It effectively degrades indigo dye. In the future, the crude enzyme solution containing laccase could be processed into dry powder for industrial production, addressing the drawbacks of current jeans whitening technologies, such as safety concerns, environmental harm, immaturity of methods, and high costs. | ||
| + | |||
| + | This new product would eliminate the need for harmful chemical bleaching agents, offering an eco-friendly and gentle treatment method. Additionally, Bpul laccase dry powder is easy to transport and store, poses no threat to the environment or human health, and could provide consumers with greater freedom to DIY their own jeans. It allows for more precise control over the bleaching process, reduces fabric damage, and improves both the sustainability and quality of jeans production, making it a promising development for the future. | ||
| + | ==References== | ||
| + | Mo Y, Lao H I, Au S W, et al. Expression, secretion and functional characterization of three laccases in E. coli[J]. Synthetic and Systems Biotechnology, 2022, 7(1): 474-480. | ||
<!-- --> | <!-- --> | ||
Latest revision as of 03:38, 30 September 2024
bpul laccase from Bacillus pumilus
bpul (Laccase from Bacillus pumilus)
Usage and Biology
First some trials of shaking flask cultivations were made with different parameters to define the best conditions for production of the His tagged CotA aus [http://www.dsmz.de/catalogues/details/culture/DSM-27.html Bacillus pumilus DSM 27 ( ATCC7061)] named BPUL. Due to inactivity of the enzyme in the cell lysate a purification method was established (using Ni-NTA-Histag resin and Syringe or ÄKTA method). The purified BPUL could be detected by SDS-PAGE (molecular weight of 58.6 kDa) as well as by MALDI-TOF. To improve the purification strategies the length of the linear elution gradient was increased up to 200 mL . The fractionated samples were also tested concerning their activity and revealed high activity. Optimal conditions for activity were identified. After measuring activity of BPUL a successful scale up was made up to 3 L and also up to 6 L that enables an intense screening afterwards. The scale up will be important for a further application.
- Allergen characterization of BBa_K863001: NOT a potential allergen
The Baltimore Biocrew 2017 team discovered that proteins generated through biobrick parts can be evaluated for allergenicity. This information is important to the people using these parts in the lab, as well as when considering using the protein for mass production, or using in the environment. The allergenicity test permits a comparison between the sequences of the biobrick parts and the identified allergen proteins enlisted in a data base.The higher the similarity between the biobricks and the proteins, the more likely the biobrick is allergenic cross-reactive. In the full-length alignments by FASTA, 30% or more amount of similarity signifies that the biobrick has a Precaution Status meaning there is a potential risk with using the part. A 50% or more amount of identity signifies that the biobrick has a Possible Allergen Status. In the sliding window of 80 amino acid segments, greater than 35% signifies similarity to allergens. The percentage of similarity implies the potential of harm biobricks’ potential negative impact to exposed populations. For more information on how to assess your own biobrick part please see the “Allergenicity Testing Protocol” in the following page http://2017.igem.org/Team:Baltimore_Bio-Crew/Experiments
For the biobrick part, BBa_K863001, there was a 25.0 % of identity match and 50.0% similarity match to the top allergen in the allergen database. This means that the biobrick part is not of potential allergen status. In 80 amino acid alignments by FASTA window, no matches found that are greater than 35% for this biobrick. This also means that there is not of potential allergen status.
Contents
- 1 Usage and Biology
- 2 Cultivation, Purification and SDS-PAGE
- 2.1 Shaking Flask Cultivation
- 2.2 3 L Fermentation E. coli KRX with BBa_K863000
- 2.3 Purification of BPUL
- 2.4 SDS-PAGE of purified BPUL
- 2.5 6 L Fermentation of E. coli KRX with BBa_K863000
- 2.6 Purification of BPUL
- 2.7 SDS-PAGE of purified BPUL
- 2.8 12 L Fermentation E. coli KRX with BBa_K863000
- 2.9 Since Regionals: Purification of BPUL
- 2.10 SDS-PAGE of protein purification
- 2.11 MALDI-TOF Analysis of BPUL
- 3 Activity analysis of BPUL
- 3.1 Since Regionals: Initial activity tests of purified fractions
- 3.2 BPUL CuCl2 concentration
- 3.3 BPUL activity depending on different ABTS concentrations
- 3.4 Impact of MeOH and acteonitrile on BPUL
- 3.5 Since Regionals: Searching for substrate saturation ofBPUL
- 3.6 Since Regionals: BPUL pH optimum
- 3.7 Since Regionals: BPUL activity at different temperatures
- 4 Substrate Analysis
- 5 Immobilization
- 6 Xiamen_City 2019's characterization
- 7 BBa_K863001 bpul laccase from Bacillus pumilus
- 8 Contribution From GEC-Beijing
Cultivation, Purification and SDS-PAGE
Shaking Flask Cultivation
The first trials to produce the CotA-laccase from [http://www.dsmz.de/catalogues/details/culture/DSM-27.html Bacillus pumilus DSM 27] (ATCC7061, named BPUL) were performed in shaking flasks with various designs (from 100 mL-1 to 1 L flasks, with and without baffles) and under different conditions. The parameters tested during the screening experiments were temperature (27 °C,30 °C and 37 °C), the concentration of chloramphenicol (20 to 170 µg mL-1), induction strategy ([http://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Autoinduction_medium autoinduction] and manual induction with 0,1 % rhamnose) and cultivation time (6 to 24 h). Furthermore it was cultivated with and without 0.25 mM CuCl2, to provide a sufficient amount of copper, which is needed for the active center of the laccase. Based on the screening experiments the best conditions for expression of BPUL were identified(see below). The addition of CuCl2 did not increase activity, so it was omitted.
- flask design: shaking flask without baffles
- medium: [http://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Autoinduction_medium autoinduction medium]
- antibiotics: 60 µg mL-1 chloramphenicol
- temperature: 37 °C
- cultivation time: 12 h
The reproducibility of the measured data and results were investigated for the shaking flask and bioreactor cultivation.
3 L Fermentation E. coli KRX with BBa_K863000
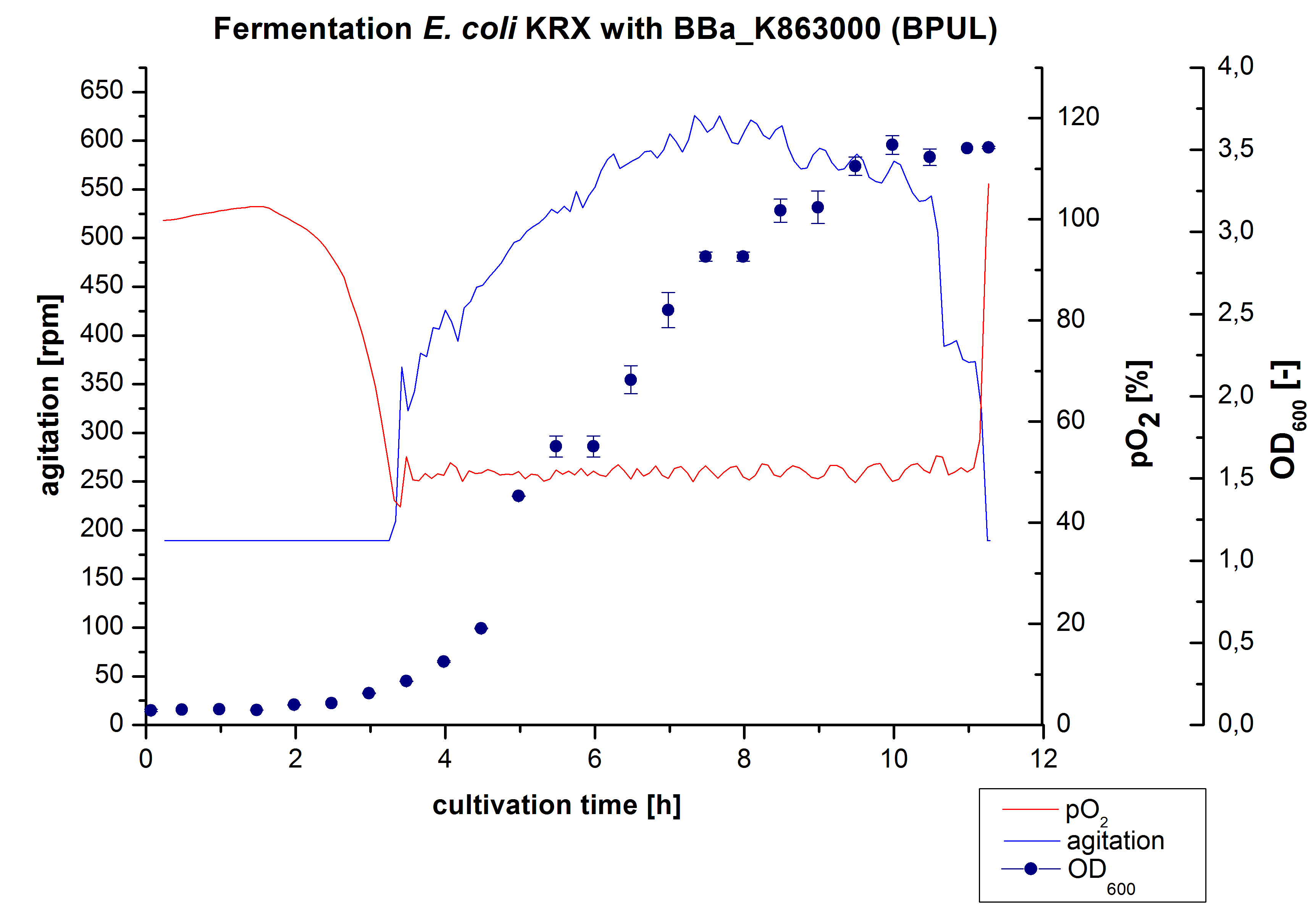
After the measurement of BPUL activity we made a scale-up and fermented E. coli KRX with BBa_K863000 in a[http://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Biostat_B_Bioreactor_.283_L.29_by_Braun Braun Biostat B] fermenter with a total volume of 3 L. Agitation speed, pO2 and OD600 were determined and illustrated in Figure 1. We got a long lag phase of 2 hours due to a relatively old preculture. The cell growth caused a decrease in pO2 and after 3 hours the value fell below 50 %, so that the agitation speed increased automatically. After 8.5 hours the deceleration phase started and therefore the agitation speed was decreased. The maximal OD600 of 3.53 was reached after 10 hours, which means a decrease in comparison to the fermentation of E. coli KRX under the same conditions (OD600,max =4.86 after 8.5 hours, time shift due to long lag phase). The cells were harvested after 11 hours.
Purification of BPUL
The harvested cells were resuspended in [http://2012.igem.org/wiki/index.php?title=Team:Bielefeld-Germany/Protocols/Materials#Buffers_for_His-Tag_affinity_chromatography Ni-NTA-equilibrationbuffer], mechanically lysed by [http://2012.igem.org/Team:Bielefeld-Germany/Protocols/Production#Mechanical_lysis_of_the_.28bio-reactor.29_cultivation homogenization] and cell debris were removed by centrifugation. The supernatant of the lysed cell paste was loaded on the Ni-NTA-column (15 mL Ni-NTA resin) with a flowrate of 1 mL min-1 cm-2. The column was washed with 10 column volumes (CV) Ni-NTA-equilibrationbuffer. The bound proteins were eluted by an increasing Ni-NTA-elutionbuffer gradient from 0 % to 100 % with a total volume of 100 mL and the elution was collected in 10 mL fractions. Due to the high UV-detection signal of the loaded samples and to simplify the illustration of the detected product peak only the UV-detection signal of the wash step and the elution are shown. A typical chromatogram of purified laccases is illustrated here. The chromatogram of the BPUL-elution is shown in Figure 2:
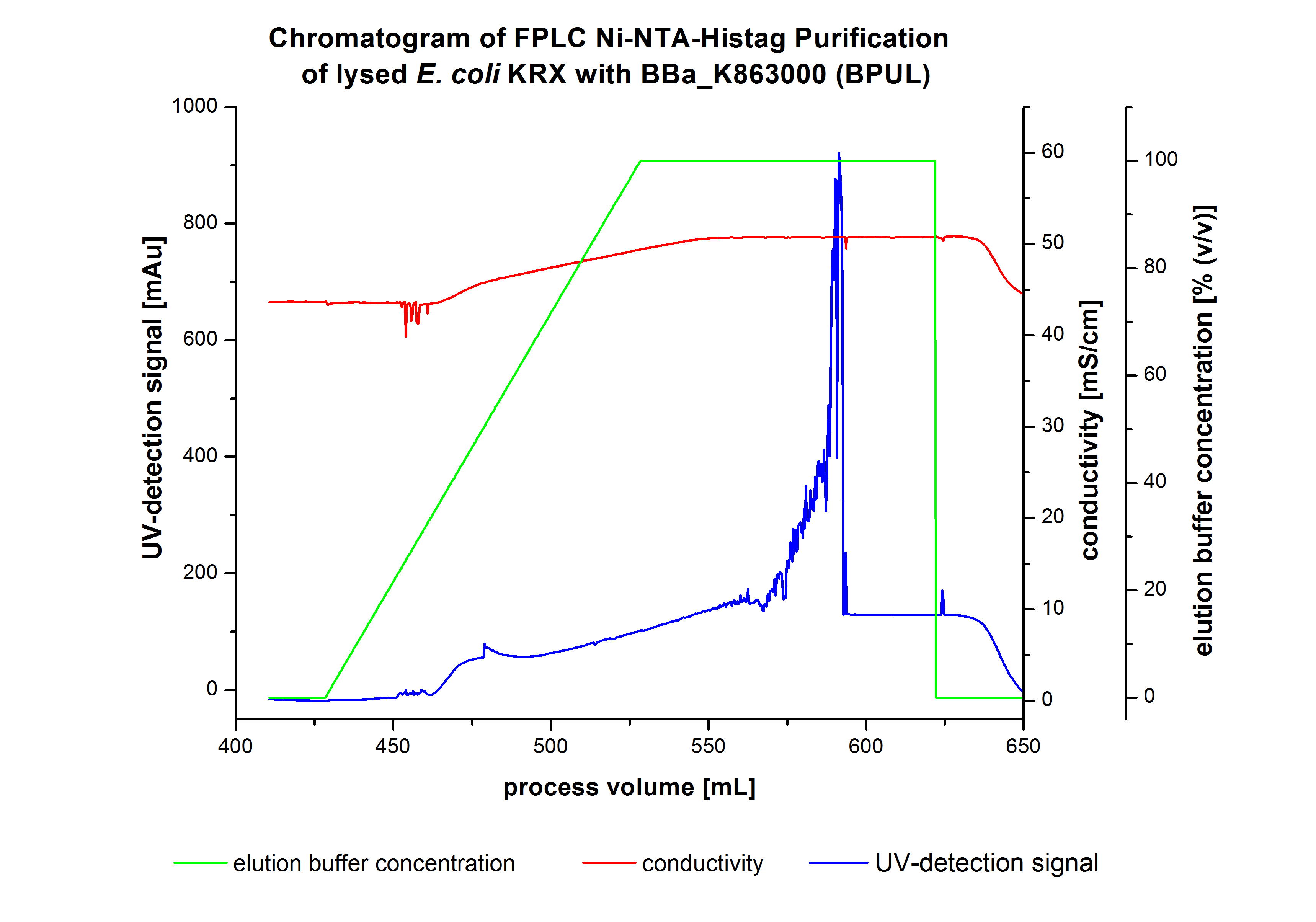
The chromatogram shows a remarkable widespread peak between the process volume of 460 mL to 480 mL with the highest UV-detection signal of 69 mAU, which can be explained by the elution of bound proteins. The corresponding fractions were analyzed by SDS-PAGE analysis. During the elution, a steady increase of the UV-signal caused by the increasing imidazol concentration during the elution gradient. Between the process volume of 550 and 580 mL there are several peaks (up to a UV-detection-signal of 980 mAU) detectable. These results are caused by an accidental detachment in front of the UV-detector. Just to be on the safe side, the corresponding fractions were analyzed by SDS-PAGE analysis. The corresponding SDS-PAGE is shown in Figure 3.
SDS-PAGE of purified BPUL
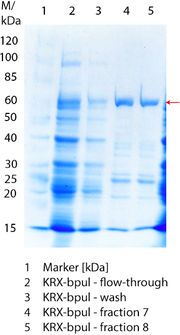
Figure 3 shows the purified ECOL including flow-through, wash and the elution fractions 7 and 8. These two fractions were chosen due to a high peak in the chromatogram. BPUL has a molecular weight of 58.6 kDA and was marked with a red arrow. The band appears in both fractions. There are also some other non-specific bands, which could not be identified. To improve the purification the elution gradient length should be longer and slower the next time.
The appearing bands were analyzed by MALDI-TOF and could be identified as CotA (BPUL).
6 L Fermentation of E. coli KRX with BBa_K863000
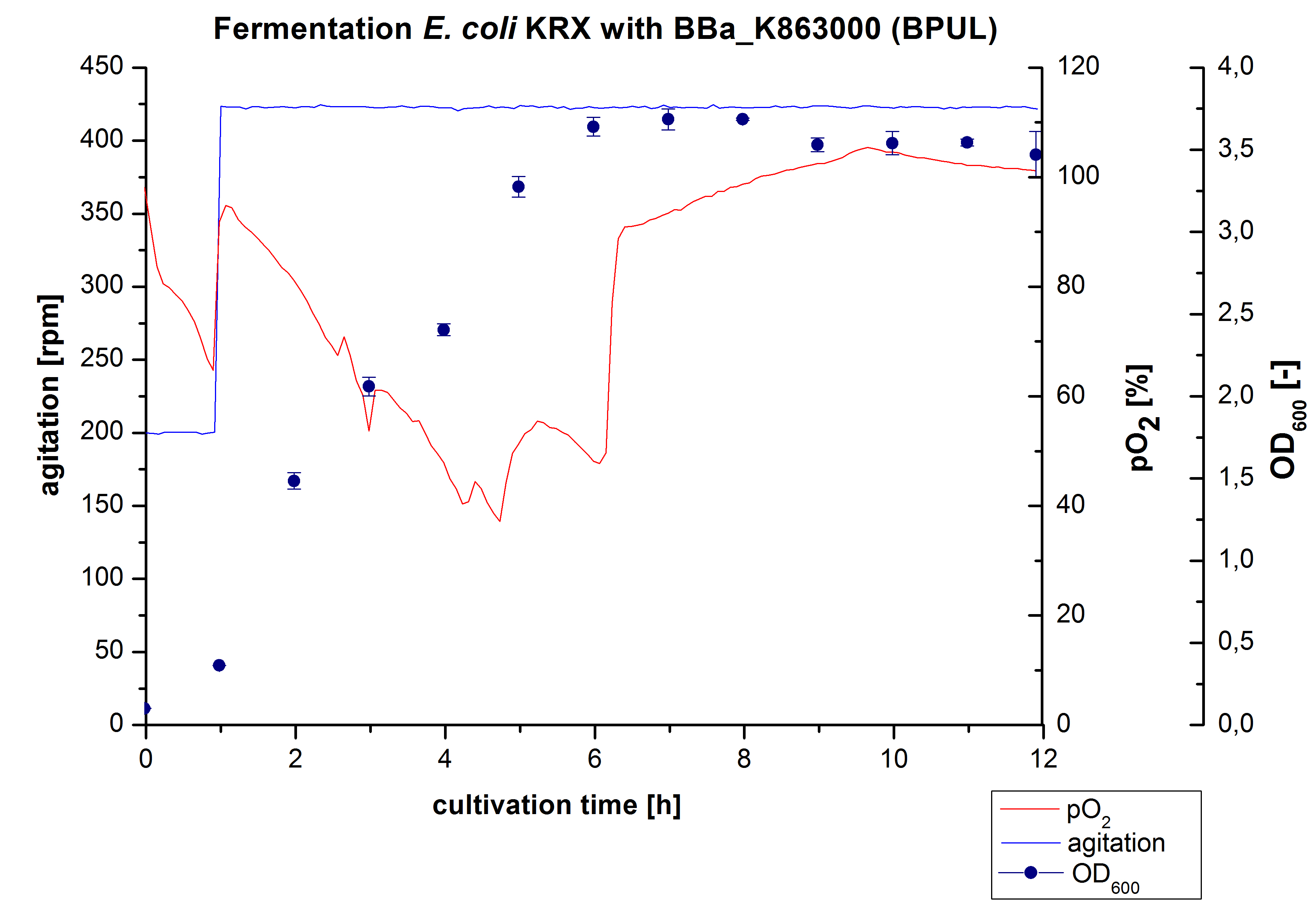
Another scale-up for E. coli KRX with BBa_K863000 was made up to a final working volume of 6 L in a Bioengineering NFL22. Agitation speed, pO2 and OD600 were determined and illustrated in Figure 4. There was no noticeable lag phase. Agitation speed was increased up to 425 rpm after one hour due to problems caused by the control panel. The pO2 decreased until a cultivation time of 4.75 hours. The increasing pO2 level indicates the beginning of the deceleration phase. There is no visible break in cell growth caused by an induction of protein expression. A maximal OD600 of 3.68 was reached after 8 hours of cultivation, which is similar to the 3 L fermentation (OD600 = 3.58 after 10 hours, time shift due to long lag phase). The cells were harvested after 12 hours.
Purification of BPUL
The harvested cells were prepared in [http://2012.igem.org/wiki/index.php?title=Team:Bielefeld-Germany/Protocols/Materials#Buffers_for_His-Tag_affinity_chromatography Ni-NTA-equilibrationbuffer], mechanically lysed by [http://2012.igem.org/Team:Bielefeld-Germany/Protocols/Production#Mechanical_lysis_of_the_.28bio-reactor.29_cultivation homogenization] and cell debris were removed by centrifugation. The supernatant of the lysed cell paste was loaded on the Ni-NTA-column (15 mL Ni-NTA resin) with a flow rate of 1 mL min-1 cm-2. The column was washed with 5 column volumes (CV) [http://2012.igem.org/wiki/index.php?title=Team:Bielefeld-Germany/Protocols/Materials#Buffers_for_His-Tag_affinity_chromatography Ni-NTA-equilibrationbuffer]. The bound proteins were eluted by an increasing elutionbuffer gradient from 0 % (equates to 20 mM imidazol) to 100 % (equates to 500 mM imidazol) with a length of 200 mL. This strategy was chosen to improve the purification by a slower increase of [http://2012.igem.org/wiki/index.php?title=Team:Bielefeld-Germany/Protocols/Materials#Buffers_for_His-Tag_affinity_chromatography Ni-NTA-elutionbuffer] concentration. The elution was collected in 10 mL fractions. Due to the high UV-detection signal of the loaded samples and to simplify the illustration of the detected product peak only the UV-detection signal of the wash step and the elution are shown. A typical chromatogram of purified laccases is illustrated here. The chromatogram of the BPUL elution is shown in Figure 5.
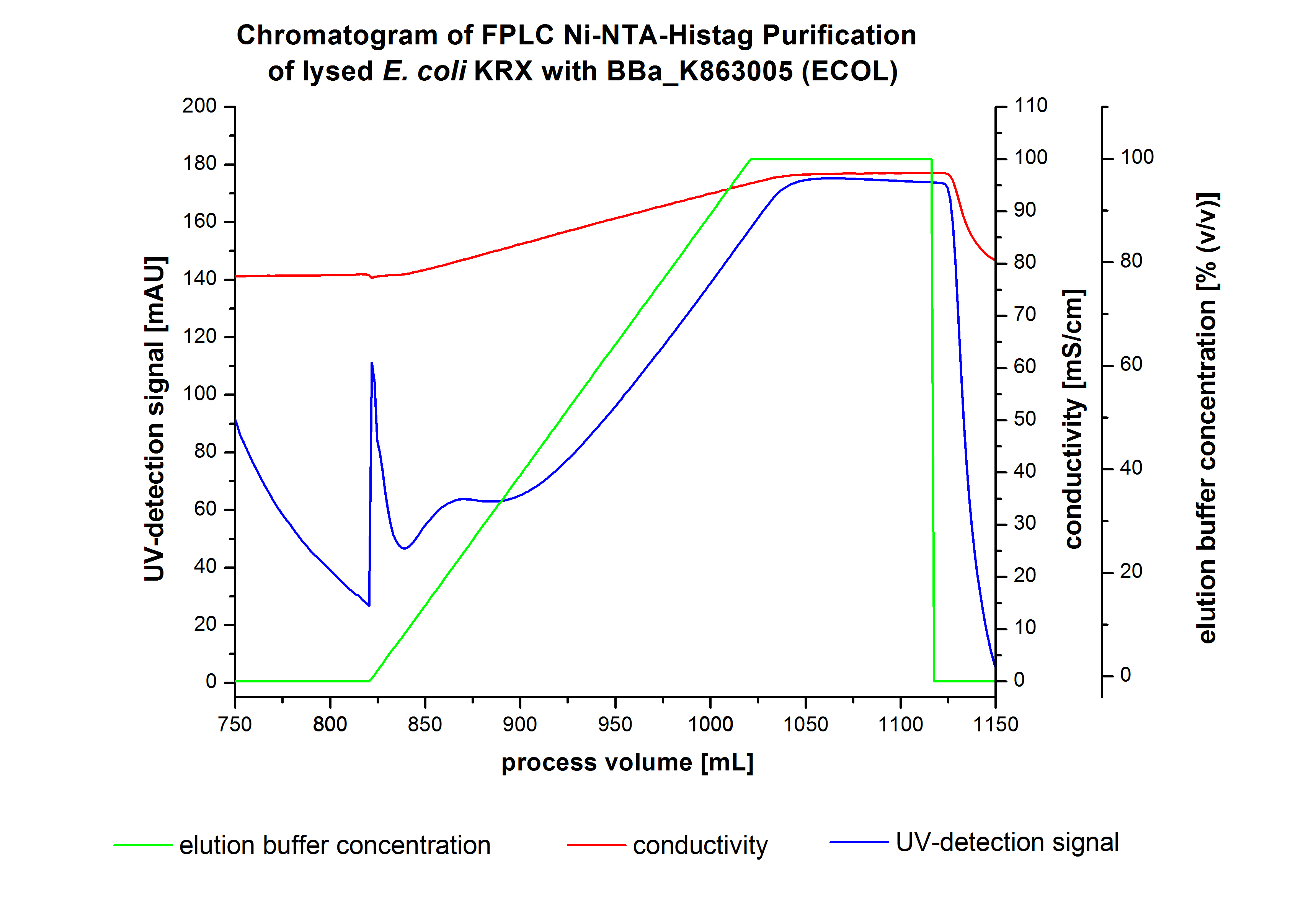
The chromatogram shows a peak at the beginning of the elution. This can be explained by pressure fluctuations upon starting the elution procedure. In between the processing volumes of 832 mL and 900 mL there is remarkable widespread peak with a UV-detection signal of 115 mAU. This peak corresponds to an elution of bound proteins at a [http://2012.igem.org/wiki/index.php?title=Team:Bielefeld-Germany/Protocols/Materials#Buffers_for_His-Tag_affinity_chromatography Ni-NTA elution buffer] concentration between 10 % and 20 % (equates to 50-100 mM imidazol). The corresponding fractions were analyzed by SDS-PAGE. The ensuing upwards trend of the UV-signal is caused by the increasing imidazol concentration during the elution gradient. Towards the end of the elution procedure there is a constant UV-detection signal, which shows, that most of the bound proteins was already eluted. Just to be on the safe side, all fractions were analyzed by SDS-PAGE to detect BPUL. The SDS-PAGE is shown in Figure 6.
SDS-PAGE of purified BPUL
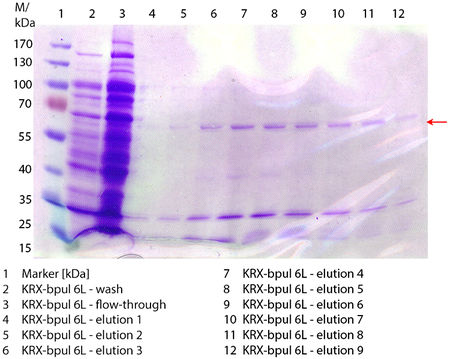
In Figure 6 the SDS-PAGE of the Ni-NTA purification of the lysed E. coli KRX culture containing BBa_K863000 is illustrated. It shows the flow-through, wash and elution fractions 1 to 9. The His-tagged BPUL has a molecular weight of 58.6 kDA and was marked with a red arrow. The band appears in all fractions from 2 to 9 with varying strength, the strongest ones in fractions 7 to 9. There are also some other non-specific bands, which could not be identified. Therefore the purification method could moreover be improved. In summary, the scale up was successful, improving protein production and purification method once again.
Furthermore the bands were analyzed by MALDI-TOF and identified as CotA (BPUL).
12 L Fermentation E. coli KRX with BBa_K863000
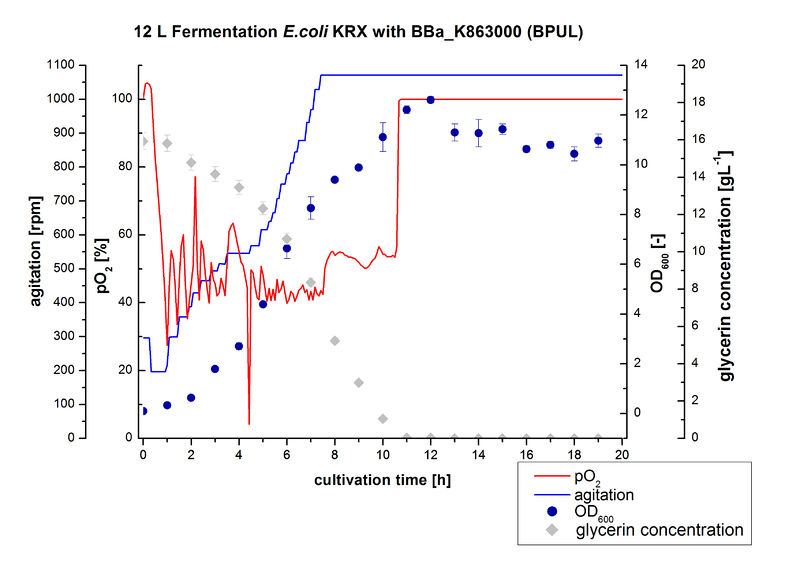
Finally another scale-up was made and E. coli KRX with BBa_K863000 was fermented in a Bioengineering NLF 22 fermenter with a total volume of 12 L to produce a high amount of the enzyme for further characterizations. This time [http://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#HSG_Autoinduction_medium HSG autoinduction medium] was used to get a higher biomass. Agitation speed, pO2 and OD600 were determined and the glycerin concentration of the samples analyzed. The data are illustrated in Figure 7. At the beginning of the cultivation, the cells were in lag phase, in which they adapt to the medium. During their growth the cells consumed glycerin as well as O2, which leads to an increase of agitation speed to hold a minimal pO2 of 50 %. After 11 hours, the glycerin was completely consumed and the pO2 increased up to 100 %, which indicates that the cells entered the stationary phase. The maximal OD600 of 12.6 was reached after 12 hours of cultivation. The cells were harvested after 19 hours of cultivation.
Since Regionals: Purification of BPUL
The harvested cells were resuspended in [http://2012.igem.org/wiki/index.php?title=Team:Bielefeld-Germany/Protocols/Materials#Buffers_for_His-Tag_affinity_chromatography Ni-NTA- equilibration buffer], mechanically disrupted by [http://2012.igem.org/Team:Bielefeld-Germany/Protocols/Production#Mechanical_lysis_of_the_.28bio-reactor.29_cultivation homogenization] and cell debris were removed by centrifugation, microfiltration as well as diafiltration to concentrate the protein concentration in the cell lysate solution. This solution of the cell lysate was loaded on the Ni-NTA column (15 mL Ni-NTA resin) with a flow rate of 1 mL min-1 cm-2. Then the column was washed with 10 column volumes (CV) [http://2012.igem.org/wiki/index.php?title=Team:Bielefeld-Germany/Protocols/Materials#Buffers_for_His-Tag_affinity_chromatography Ni-NTA equilibration buffer]. The bound proteins were eluted by an increasing [http://2012.igem.org/wiki/index.php?title=Team:Bielefeld-Germany/Protocols/Materials#Buffers_for_His-Tag_affinity_chromatography Ni-NTA elution buffer] step elution from 5 % (equates to 25 mM imidazol) with a length of 50 mL, to 50 % (equates to 250 mM imidazol) with a length of 70 mL, to 80 % (equates to 400 mM imidazol) and finally to 100 % (equates to 500 mM imidazol) with a length of 100 mL. This strategy was chosen to improve the purification caused by a step by step increasing Ni-NTA-elution buffer concentration. The elution was collected in 10 mL fractions. A typical chromatogram of purified laccases is illustrated here. Unfortunately, the data of this procedure are not available due to a computer crash after the purification step. All Fractions were analysed to detect BPUL.
SDS-PAGE of protein purification
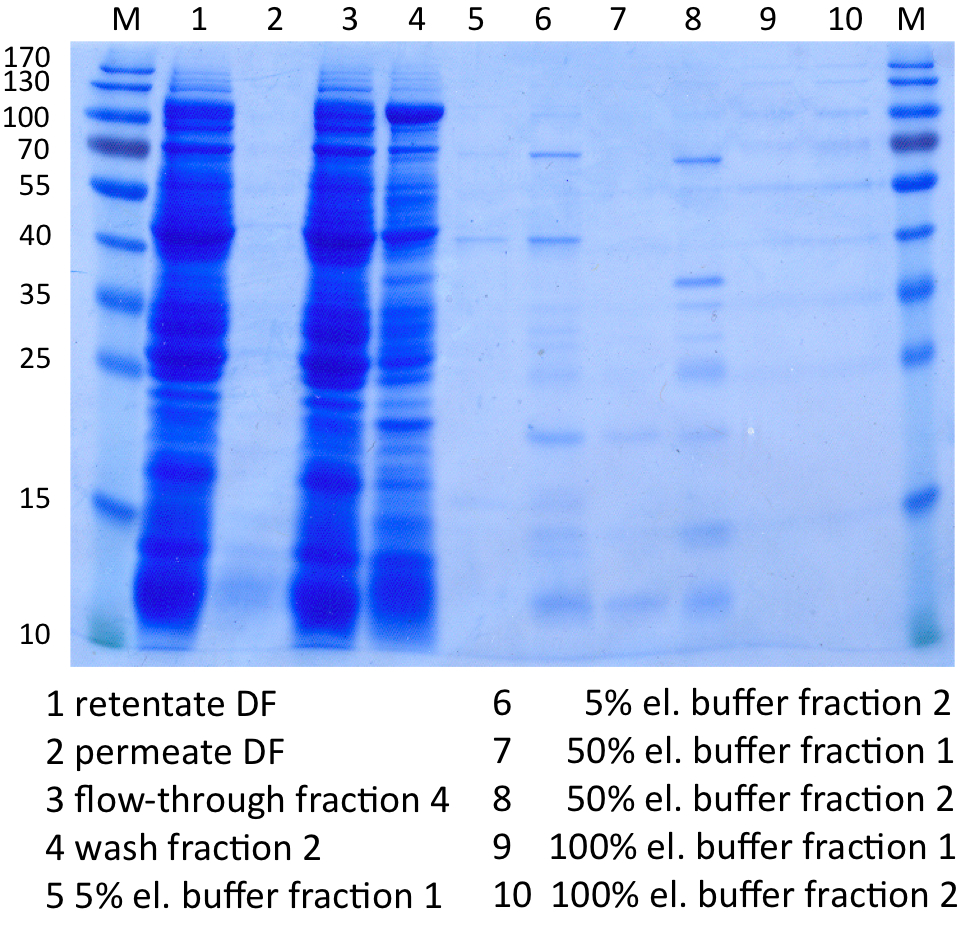
In Figure 8 the SDS-PAGE of the Ni-NTA purification of the lysed E.coli KRX culture containing BBa_K863000 is illustrated. It shows the permeate and retentate of microfiltration and diafiltration respectively, several fractions of flow-through, wash and the elutions with different buffer concentrations respectively. The selected samples were taken where peaks were seen in the chromatogram. The His-tagged BPUL has a molecular weight of 58.6 kDa. BPUL could not be attributed exactly to any band. There are some other non-specific bands, wich could not be identified because the MALDI was broken-down for the last two weeks.
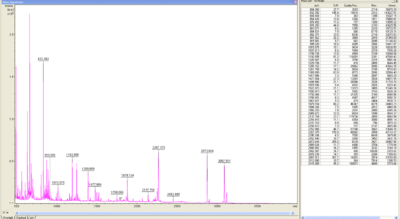
MALDI-TOF Analysis of BPUL
The E. coli laccase was identified using the following software
- FlexControl
- Flexanalysis and
- Biotools
from Brunker Daltronics.
The peptid mass fingerprints were compared with the measured fingerprint gotten from the Maldi. With a sequence coverage of 21,9% BPUL was identified.
In Figure 9 and 10 the chromatogram of the peptide mass fingerprint and the single masses is shown.
Activity analysis of BPUL
Since Regionals: Initial activity tests of purified fractions
After the Regional Jamboree in Amsterdam further BPUL was produced. The most comprising fractions of the purification were analyzed for [http://2012.igem.org/Team:Bielefeld-Germany/Amsterdam/Labjournal#Tuesday_October_16th/ protein content] (10/16), re-buffered into deionized H2O and incubated in 0.4 mM CuCl2. Again, the [http://2012.igem.org/Team:Bielefeld-Germany/Amsterdam/Labjournal#Wednesday_October_17th/ protein content] (10/17) of each fraction was determined because of the loss of proteins through re-buffering. Initial activity tests were done in Britton-Robinson buffer with 0.1 mM ABTS. The protein content of each fraction was adjusted for comparison of the resulting activity (see Fig. 11).
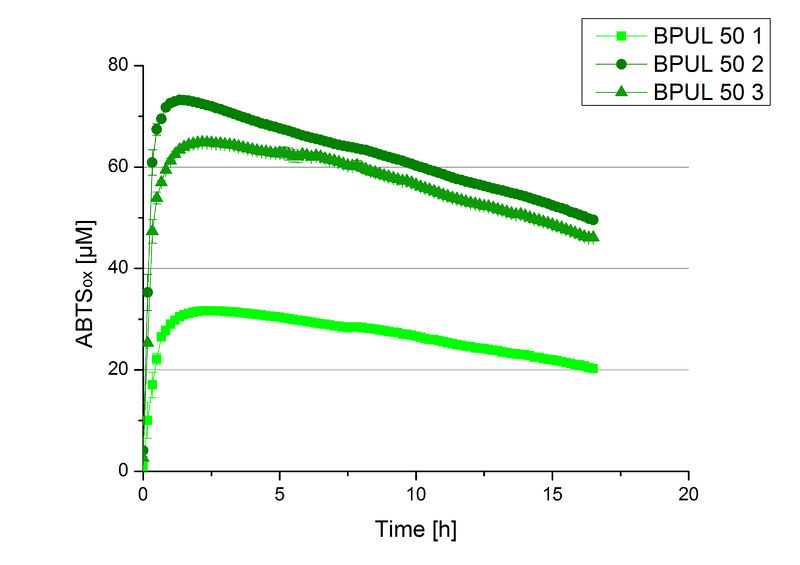
Fraction 50% 2 showed the highest activity. The first number indicates the percentage of used elution buffer, whereas the second number stands for the fraction number of this elution. The saturation was reached at ~1 h. For comparison it was stated that this fraction contains 90 % laccase and therefore the BPUL concentration is 25,1 µg mL-1.
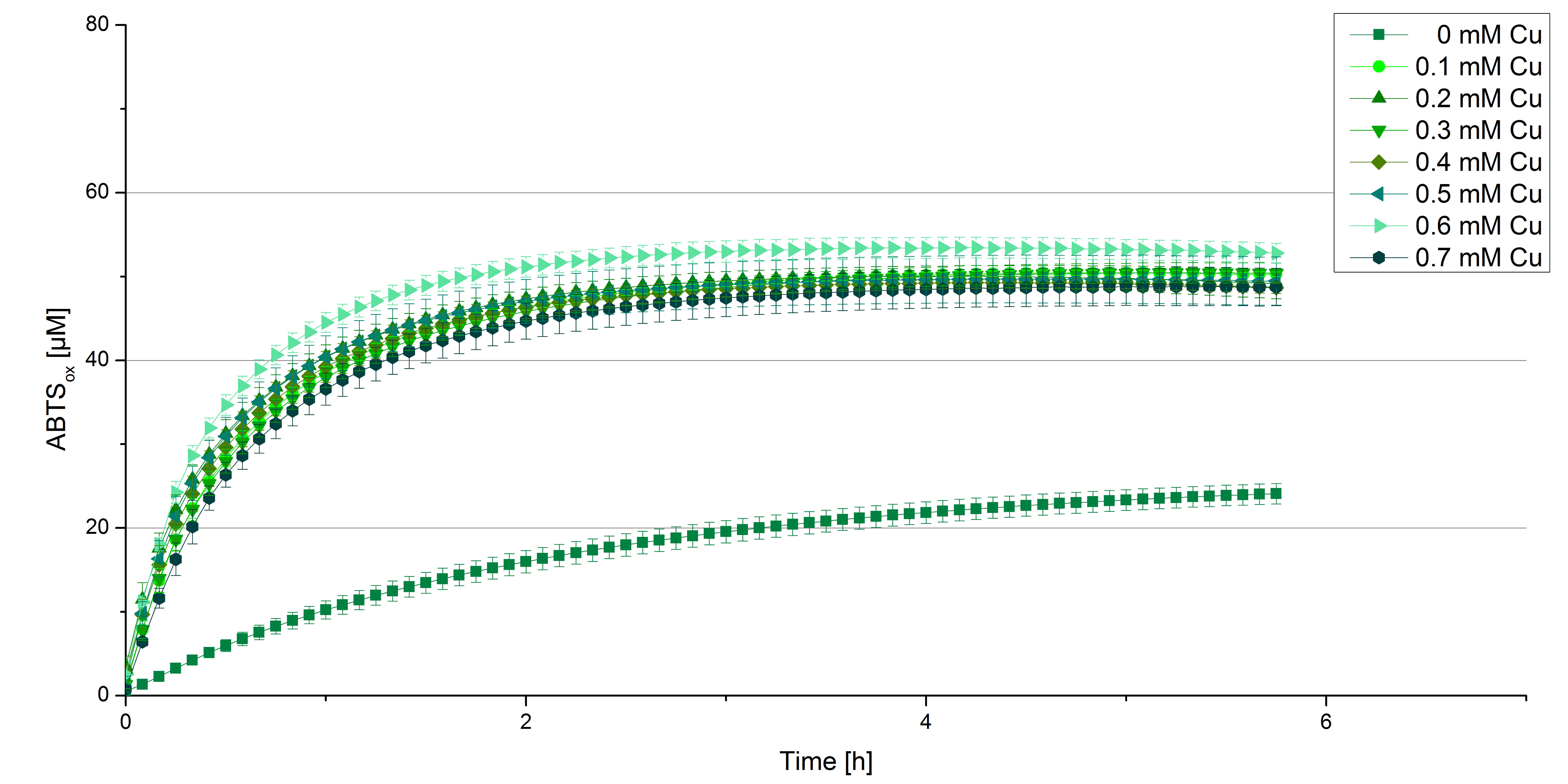
BPUL CuCl2 concentration
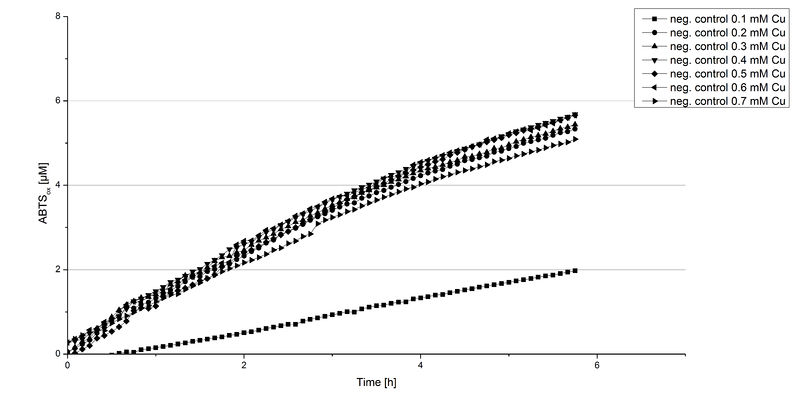
Another test of BPUL was done to survey the best CuCl2 concentration for the activity of the purified BPUL laccase. 0.03 mg mL-1 of protein were incubated with different CuCl2 concentrations ranging from 0 to 0.7 mM CuCl2. Activity tests were performed with the incubated samples, 0.1 mM ABTS and 100 mM sodium actetate buffer (pH 5) to a final volume of 200 µL. The reactivity was measured at 420 nm, 25°C and over a time period of 5 hours. As expected the saturation takes place after 3 hours (see Figure 12). The differences in the activity of BPUL laccases incubated in different CuCl2 differ minimal. The highest activity of BPUL laccase is observed after incubation with 0.6 mM CuCl2 (52% of added ABTS). With a higher concentration of 0.7 mM CuCl2 the activity seems to be reduced (only 48% ABTS got oxidized). This leads to the assumption that CuCl2 supports the BPUL laccase reactivity but concentrations exceeding this value of CuCl2 may have a negative impact on the ability of oxidizing ABTS. This fits the expectations as laccases are copper reliant enzymes and gain their activity through the incorporation of copper. Additionally negative controls were done using the tested concentrations of CuCl2 without applying laccase to detect the oxidization of ABTS through copper (see Figure 13). The more CuCl2 was present, the more ABTS was oxidzied after 5 hours. Still the maximal change accounts only for ~6% oxidized ABTS after 5 hours.
In relation to apply the laccase in waste water treatment plants it is beneficial knowing, that small amounts of CuCl2 are enough to activate the enzyme. Still it is expensive to be reliant on CuCl2 and a potential risk using heavy metals for waste water purifcation.
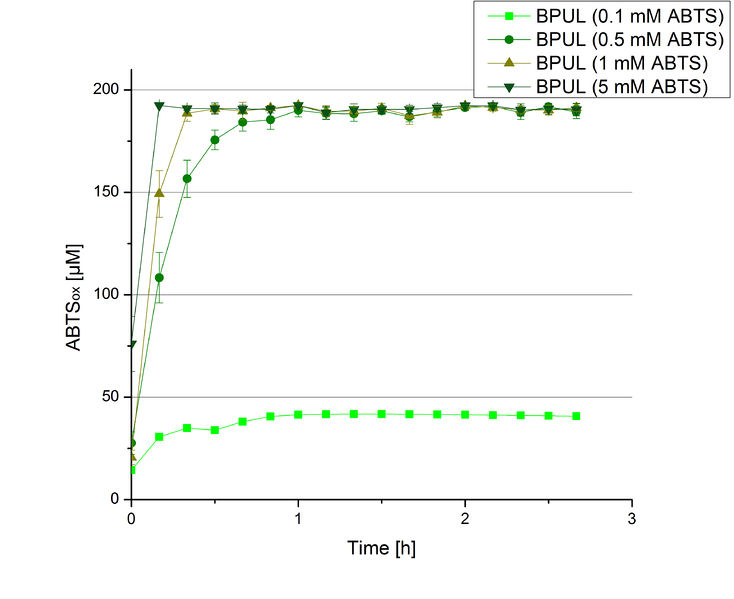
BPUL activity depending on different ABTS concentrations
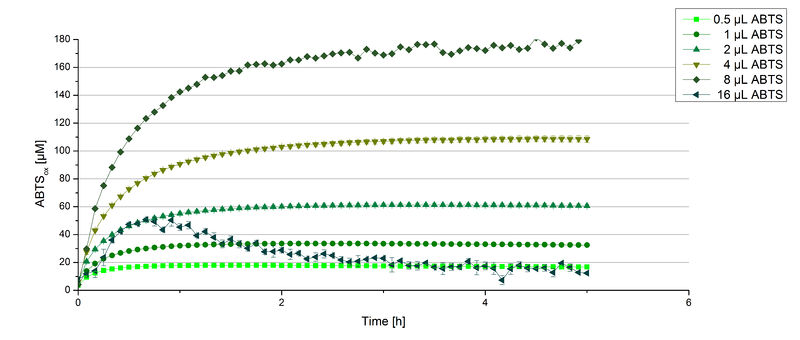
Furthermore, BPUL laccase were tested using different amounts of ABTS to calculate KM and Kcat values. The same measurement setup as described above was used only with different amounts of ABTS. As anticipated, the amount of oxidized ABTS increased in dependence of the amount of ABTS used (Figure 14). Especially using 16 µL showed an increase in the activity until 1 hour (reaching 50 µM ABTSox), but the amount of oxidized ABTS decreased afterward.
Impact of MeOH and acteonitrile on BPUL
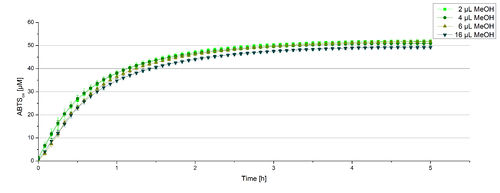
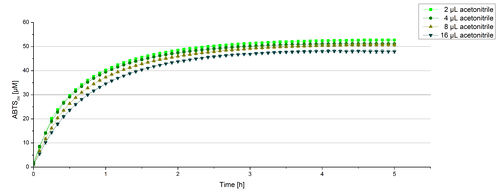
For substrate analytic tests the influence of MeOH and acetonitrile on BPUL laccases had to be determined, because substrates have to be dissolved in these reagents. The experiment setup included 0.03 mg mL-1 BPUL laccase, different amounts of MeOH (Figure 15) or acteonitrile (Figure 16), 0.1 mM ABTS and 100 mM sodium actetate buffer to a final volume of 200 µL. The observed reactivity of BPUL in regard of oxidizing ABTS did not reveal a huge decrease. The less MeOH or acetonitrile was used, the higher was the amount of oxidized ABTS after 3 hours. An application of 16 µL MeOH or acetonitrile led to a decrease of maximal 10% oxidized ABTS compared to 2 µL MeOH or acetonitrile. Negative controls are shown in [http://2012.igem.org/Team:Bielefeld-Germany/Results/coli#Impact_of_MeOH_and_acetonitrile_on_ECOL Figure 20 and 21] of the ECOL laccase. MeOH and acetonitril are able to oxidize ABTS. After 5 hours at 25°C ~15 µM ABTS get oxidized through MeOH or acetonitrile, but samples with BPUL laccase show a distinct higher activity of 50 µM ABTSox.
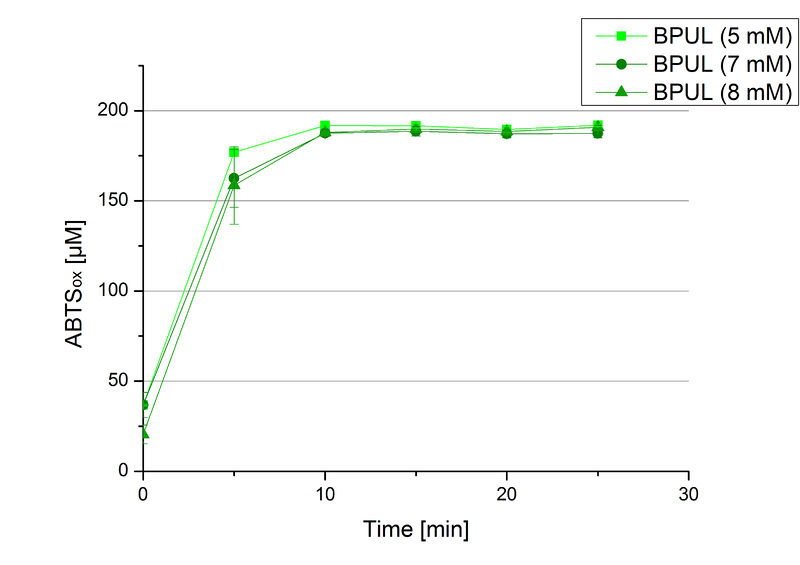
Since Regionals: Searching for substrate saturation ofBPUL
In order to find the substrate saturation, laccase activity was measured with ABTS concentrations ranging from 0.1 mM to 8 mM. 616 ng BPUL were used for measurements with ABTS concentrations of 0.1 mM to 5 mM, 308 ng BPUL were used for measurements with ABTS concentrations of 5 mM to 8 mM. Measurements were done in Britton-Robinson buffer (pH 5) at 25 °C for 30 minutes taking the OD420 every 5 minutes. Comparing the graphs in Figure 17 and Figure 18, both show a comprising substrate saturation with 5 mM ABTS. Higher concentrations of ABTS than 5 mM did not show any other effects on the activity of BPUL. For all following BPUL activity measurements after the Regional Jamborees in Amsterdam a concentration of 5 mM ABTS was applied.
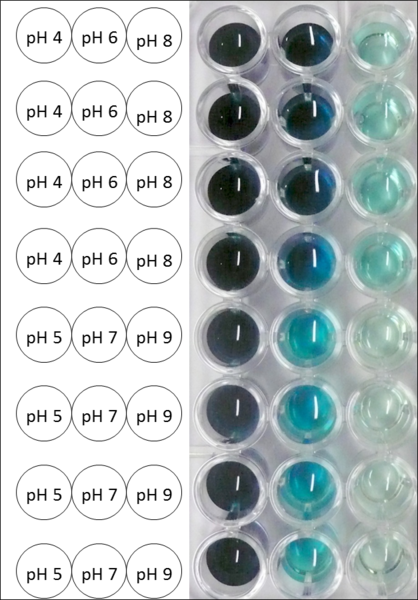
Since Regionals: BPUL pH optimum
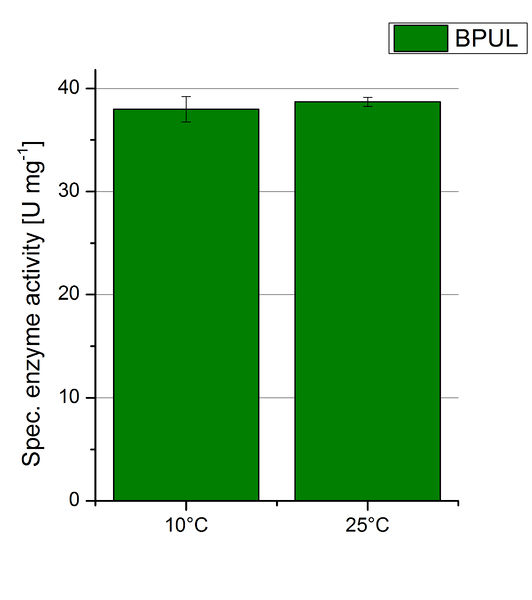
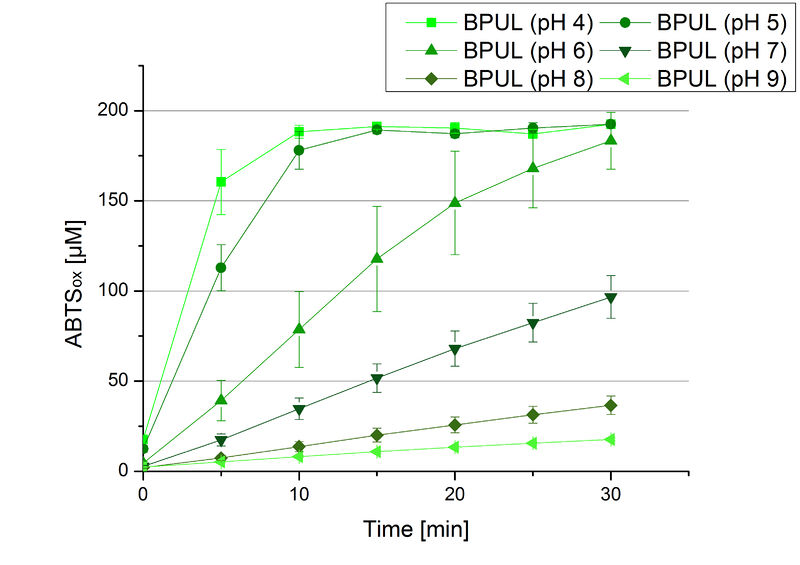
The pH of the medium containing the enzyme is of high importance for its activity. The pH optima of BPUL are pH 4 and pH 5. This is the result of activity measurements using Britton-Robinson buffer with differently adjusted pHs. BPUL was re-buffered into H2O and incubated with 0.4 mM CuCl2. The range from pH 4 to pH 9 was tested under substrate saturation at 25 °C for 30 minutes. At pH 4 and pH 5 ABTS got oxidized the fastest (see Fig. 19 and Fig. 20). At higher pHs than pH 5, the activity of BPUL was decreased considerably. The resulting Units mg-1 support the observed data (see Fig. 21). At pH 4 and pH 5 BPUL showed a specific enzyme activity of ~37 U mg-1. The higher the pH, the less U mg-1 could be calculated for BPUL. At pH 7 1/3 of the activity decreased, but still BPUL was active at this pH allowing an application of this laccase in a waste water treatment plant where the average pH is a pH of 6.9.
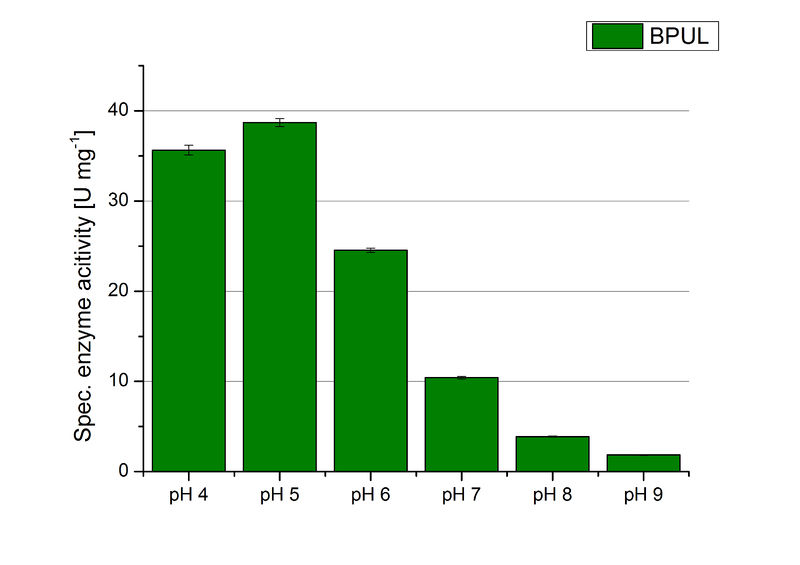
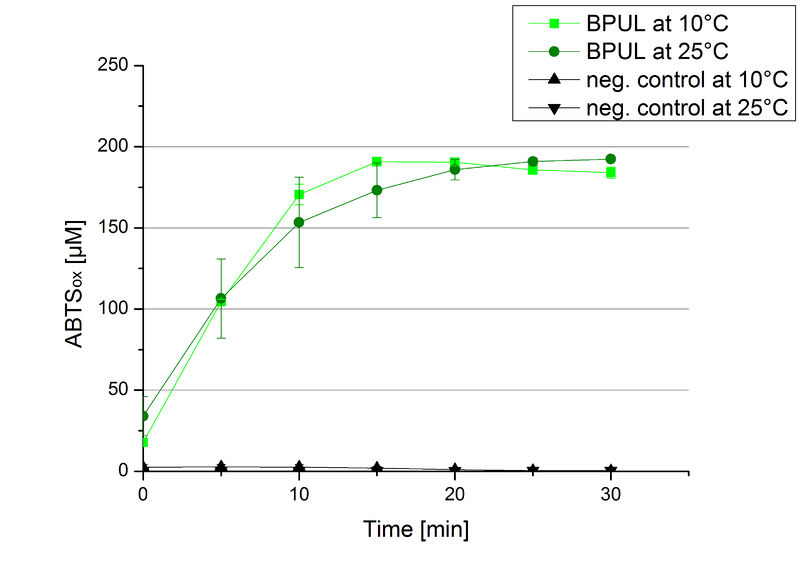
Since Regionals: BPUL activity at different temperatures
To investigate the activity of BPUL at temperatures that will apply at a waste water treatment plant throughout the year, activity tests were performed at 10 °C and 25 °C as described above. The obtained results reveal a comparable activity of BPUL at high and low temperatures (see Fig. 22). The measurements were conducted for 30 minutes until saturation initiated. Both samples reached saturation after 15-20 minutes. The obtained results were used to calculate the specific enzyme activity which was in both cases at about 37 U mg-1 (see Fig. 23). The negative control without BPUL laccase but 0.4 mM CuCl2 at 10 °C and 25 °C show a negligible oxidation of ABTS. The observed activity at both conditions was good news for the possible application in waste water treatment plants where the temperature differs from 8.1 °C to 20.8 °C.
Substrate Analysis
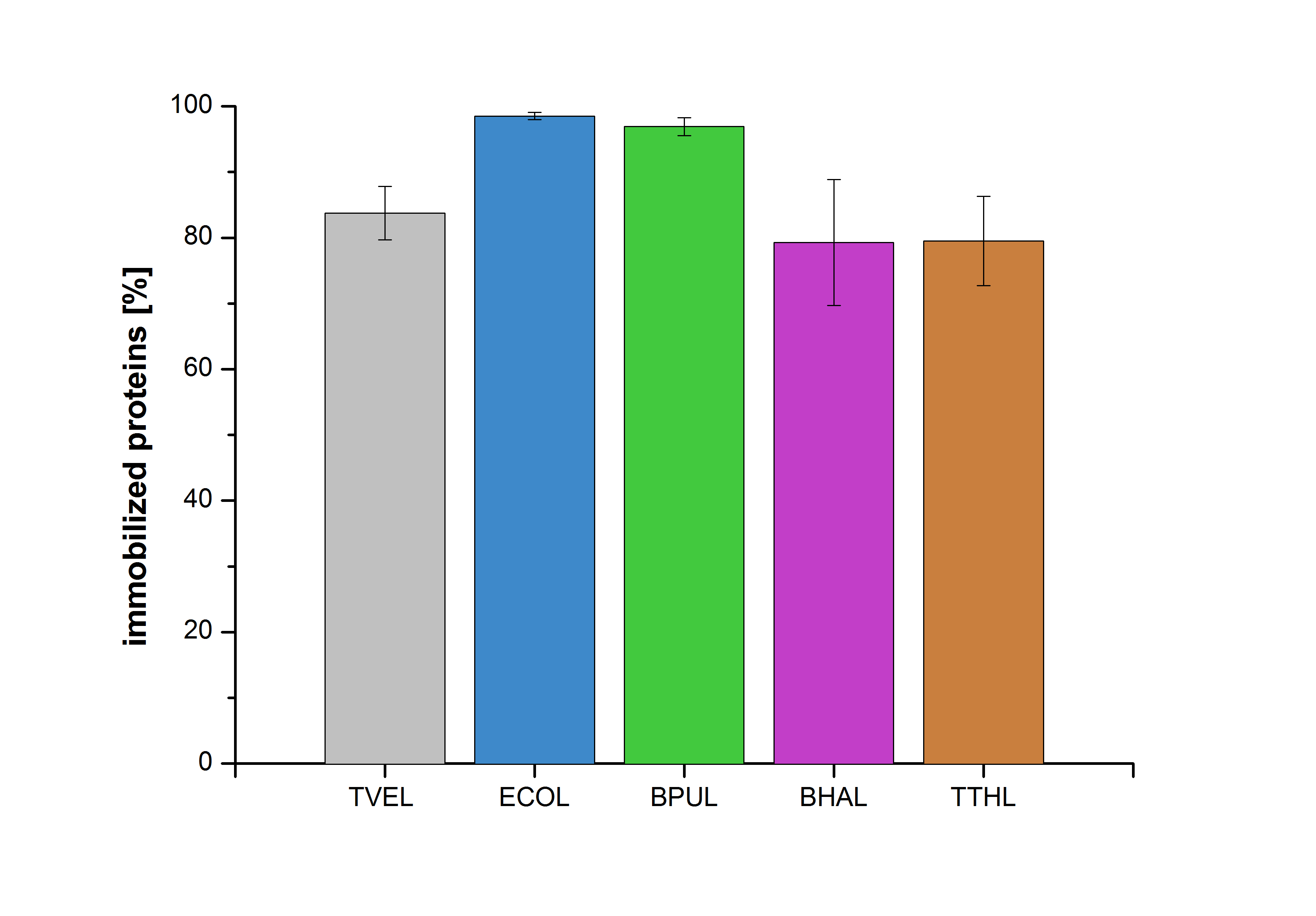
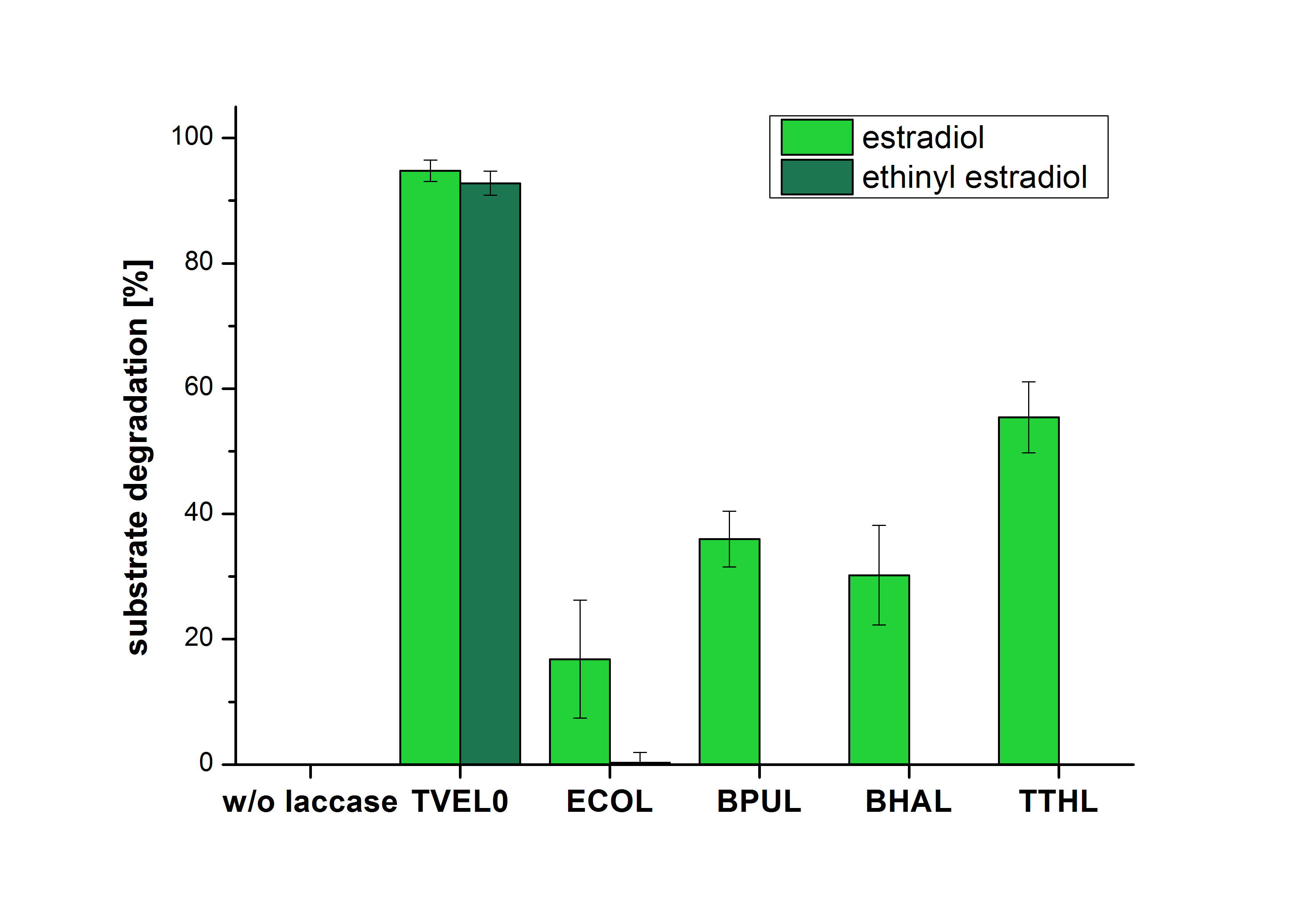
The measurements were made to test if the produced laccases were able to degrade different hormones. Therefore the produced laccases were inserted in the same concentrations (3 µg mL-1) to the different measurement approaches. To work with the correct pH value (which were measured by the Team Activity Test) Britton Robinson buffer at pH 5 was used for all measurements. The initial substrate concentration was 5 µg mL-1. The results of the reactions without ABTS are shown in Figure 24. On the Y-axis the percentages of degraded estradiol (blue) and ethinyl estradiol (red) are indicated. The X-axis displays the different tested laccases. The degradation was measured at t0 and after five hours of incubation at 30 °C. The negative control was the substrate in Britton Robinson buffer and showed no degradation of the substrates. The bought laccase TVEL0 which is used as positive control is able to degrade 94.7 % estradiol and 92.7 % ethinyl estradiol. The laccase BPUL (from Bacillus pumilus) degraded 35.9 % of used estradiol after five hours. ECOL was able to degrade 16.8 % estradiol. BHAL degraded 30.2 % estradiol. The best results were determined with TTHL (laccase from Thermus thermophilus). Here the percentage of degradation amounted 55.4 %.
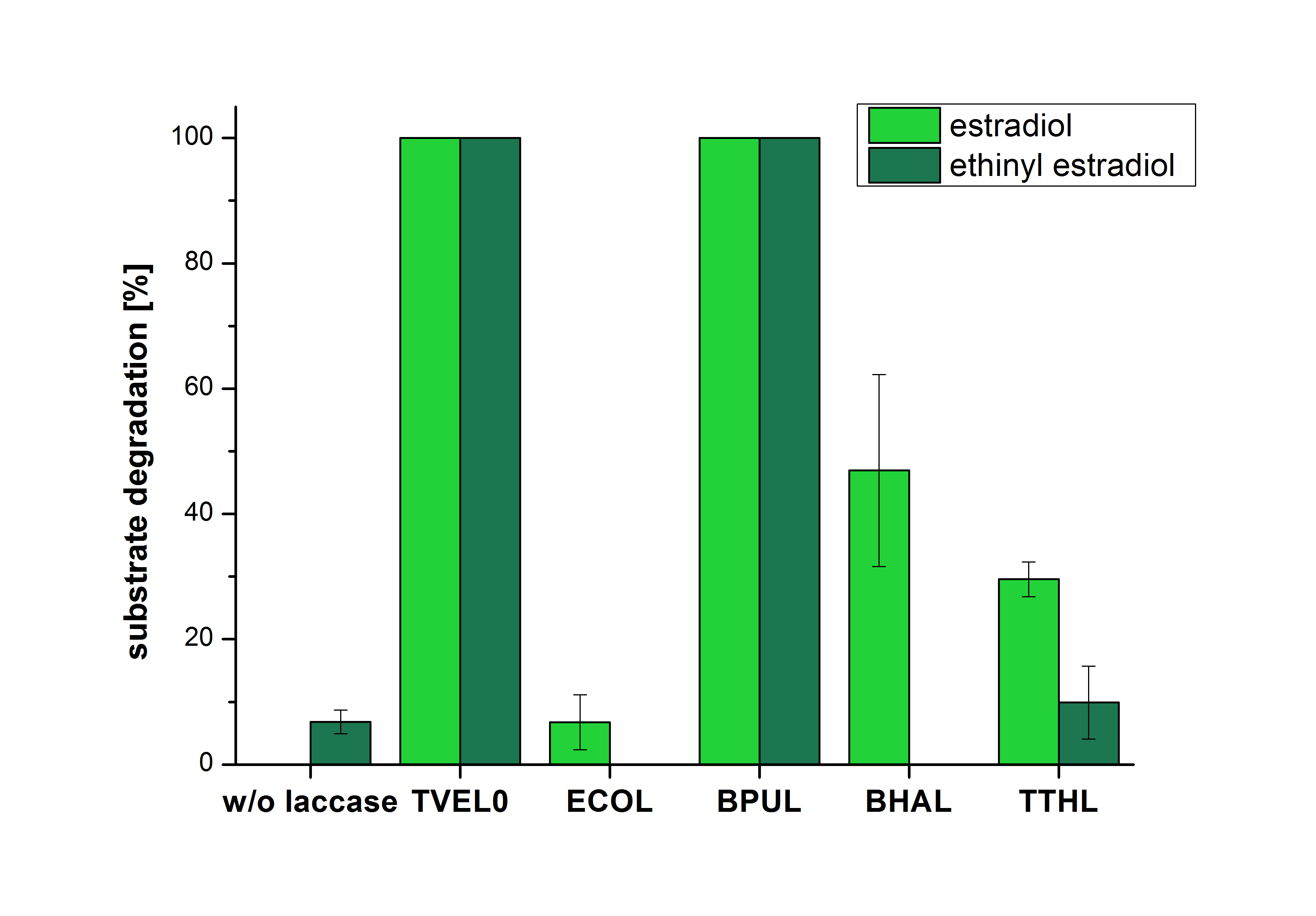
The results of the reactions of the laccases with addition of ABTS are shown in Figure 25. The experimental set ups were the same as the reaction approach without ABTS described above. The X-axis displays the different tested laccases. On the Y-axis the percentages of degraded estradiol (blue) and ethinyl estradiol (red) are shown. The degradation was measured at t0 and after five hours of incubation at 20 °C. The negative control showed no degradation of estradiol. 6.8 % of ethinyl estradiol was decayed. The positive control TVEL0 is able to degrade 100 % estradiol and ethinyl estradiol. The laccase BPUL (from Bacillus pumilus) degraded 46.9 % of used estradiol after ten minutes incubation. ECOL was able to degrade 6.7 % estradiol. BHAL degraded 46.9 % estradiol. With TTHL (laccase from Thermus thermophilus)a degradation 29.5 % were determined.
Immobilization
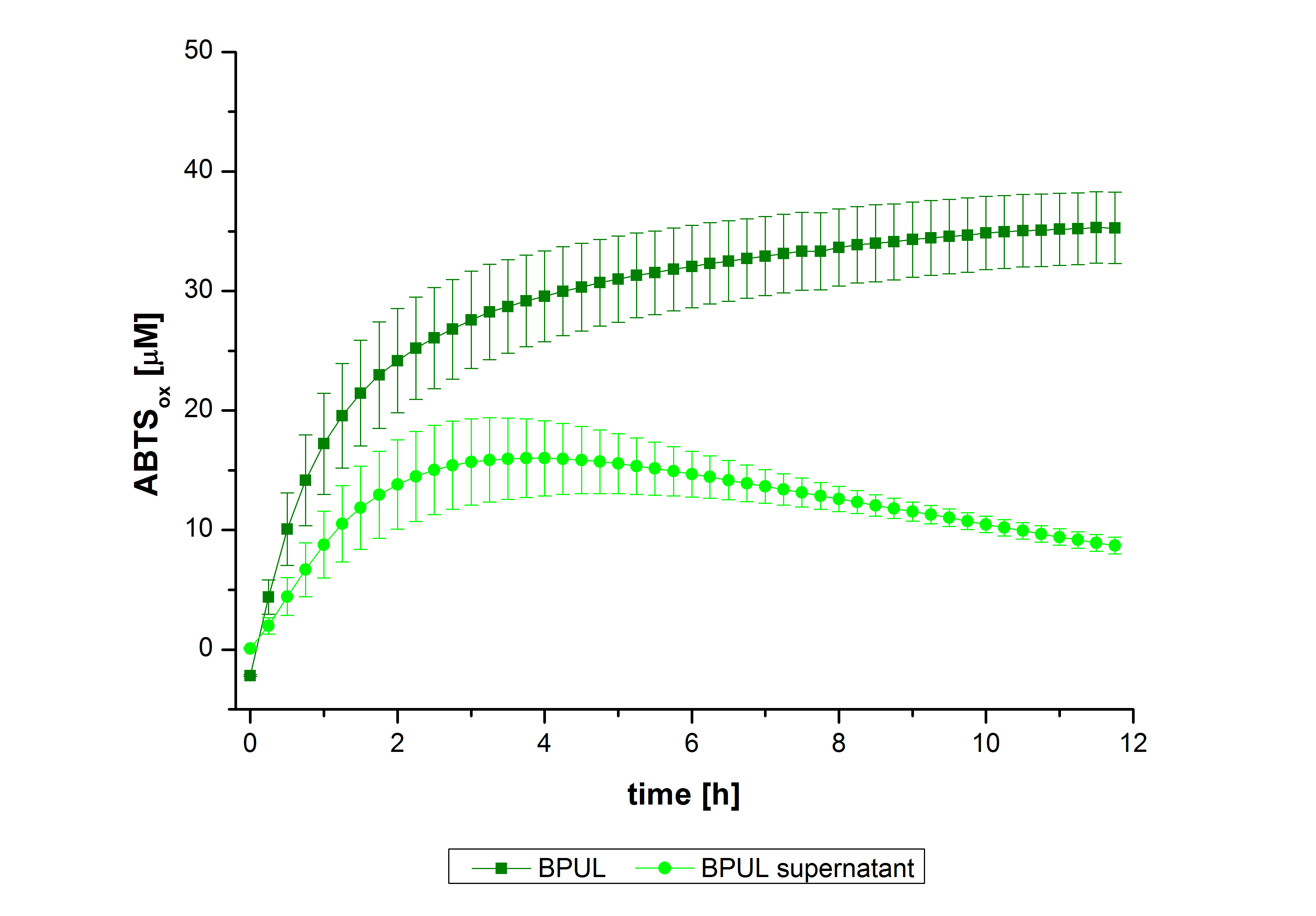
Figure 26 shows the percentage of laccases bound after incubation with CPC-beads, relative to the original concentration. The concentration of laccases in the supernatant after incubation was measured using Roti®-Nanoquant. The results showed that only 3% of BPUL laccases was still present in the supernatant. This illustrates that BPUL was successfully immobilized on the CPC-beads.
In figure 27, the enzymatic activity of BPUL in the supernatant is compared to the activity of nontreated BPUL. Although an activity can already be detected in the supernatant, this activity is low compared to the original.
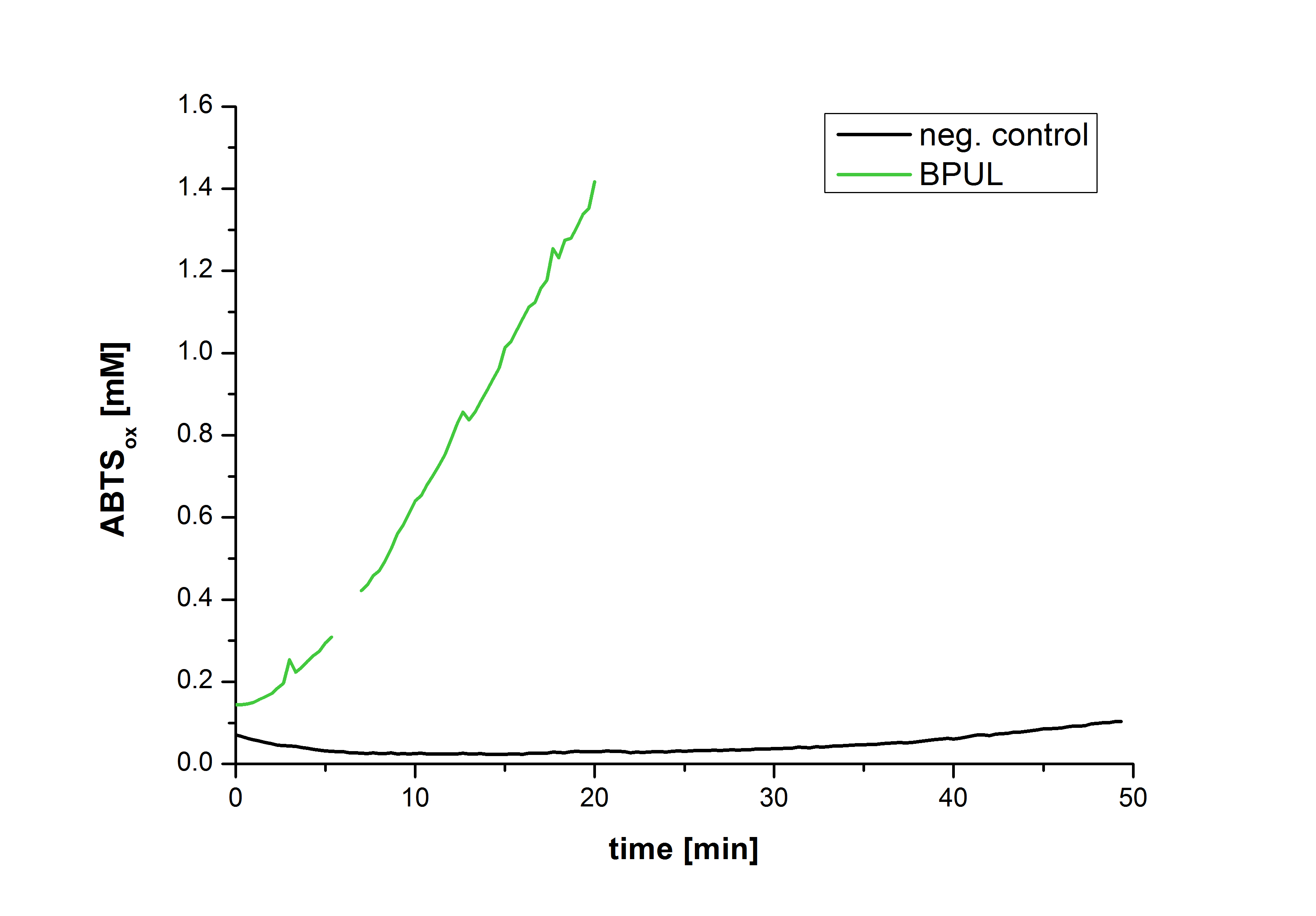
Figure 28 shows the illustration of ABTS oxidation by BPUL with time compared to the negative control. The increase in ABTS oxidized proves laccase activity even if a direct comparison with the original and not immobilized laccase solution was not possible due to the very high activity of BPUL, which could not be measured properly.
Allergen characterization of BBa_K863001
The Baltimore Biocrew 2017 team discovered that proteins generated through biobrick parts can be evaluated for allergenicity. This information is important to the people using these parts in the lab, as well as when considering using the protein for mass production, or using in the environment. The allergenicity test permits a comparison between the sequences of the biobrick parts and the identified allergen proteins enlisted in a data base.The higher the similarity between the biobricks and the proteins, the more likely the biobrick is allergenic cross-reactive. In the full-length alignments by FASTA, 30% or more amount of similarity signifies that the biobrick has a Precaution Status meaning there is a potential risk with using the part. A 50% or more amount of identity signifies that the biobrick has a Possible Allergen Status. In the sliding window of 80 amino acid segments, greater than 35% signifies similarity to allergens. The percentage of similarity implies the potential of harm biobricks’ potential negative impact to exposed populations. For more information on how to assess your own biobrick part please see the “Allergenicity Testing Protocol” in the following page http://2017.igem.org/Team:Baltimore_Bio-Crew/Experiments
For the biobrick part, BBa_K863001, there was a 25.0 % of identity match and 50.0% similarity match to the top allergen in the allergen database. This means that the biobrick part is not of potential allergen status. In 80 amino acid alignments by FASTA window, no matches found that are greater than 35% for this biobrick. This also means that there is not of potential allergen status.
Xiamen_City 2019's characterization
BBa_K863001 bpul laccase from Bacillus pumilus
(1) Transformation of Plasmid DNA
Material: Plasmid (pSB1C3-J23100-B0034-bpul-ter)
1. Take out E. coli competent cell (MG1655, DH10B, BL21(DE3)) from the refrigerator
2. Put the plasmid on ice and put it in a constant temperature water bath (42 Celsius) for 90 s
3. Use the needle tip to take a small amount of plasmid, add it to the tube with 300 μL medium, and put it onto the plate with solid media.
4. Pick the colonies in LB media and put it in shaker at 37℃
(2) Protein Electrophoresis
Material: MG1655 bpul, DH10B bpul, BL21(DE3) bpul, loading solution (6 μL), deionized water (1000 μL), marker solution, Coomassie brilliant blue dye.
1. Add 24 μL MG1655 DH10B bpul, BL21(DE3) bpul to 1.5mL tube respectively, and then add 6 μL loading.
2. Add 500 μL deionized water and centrifuge the tubes respectively.
3. Add another 500 μL deionized water, blow and suspend the solution.
4. Put the tubes 99℃ water for 15 minutes.
5. After heating, centrifuge the tubes for 5min.
6. Take and install the gel, add the electrophoresis buffer to the sample hole, and check if there is any leakage. Add marker, to the first and last sample holes, add MG1655 bpul, DH10B bpul, BL21(DE3) bpul to the other sample holes, respectively.
7. Connect the electrophoresis device to the power supply. Connect the positive electrode to the tank and the negative electrode to the slot for electrophoresis. The voltage is adjusted to 160V.
8. Turn off the power supply and disconnect the electrode until the bromophenol blue reaches the bottom of the release adhesive. Remove the glass sheet from the electrophoresis device and then remove the gel.
9. Soak the gel in Coomassie brilliant blue dye and dye it in a horizontal shaking bed for 15 min.
Observe the protein bands
(3) HPLC
Two colonies (DH10B bpul) were picked and added with 5000 um of m-phenylenediamine, and the left side in the figure was a spectrum with nothing added. The middle was added with m-phenylenediamine for one day, and the right was after adding m-phenylenediamine for three days, the content of S-1 and T-5 was found to decrease when the content was detected.
The figure shows the peak area of m-phenylenediamine that detected for m-phenylenediamine
Contribution From GEC-Beijing
Description
We modified the existing part accase gene BBa_K863001 using surface display technology, resulting in a new part INP-Bpul (BBa_K5458001). Considering the unsafe and time-consuming process of jeans bleaching technology currently used in the market, we designed to use laccase to degrade the indigo dye in jeans to achieve more precise control of the degree of jeans bleaching and reduce damage to the fabric. We chose the laccase gene Bpul from Bacillus pumilus. Laccase has many functions, such as decolorizing textiles, removing harmful substances from wastewater, and degrading many industrial pollutants.
Usage and Biology
We first selected and codon-optimized the laccase gene Bpul (BBa_K863001) from Bacillus pumilus to ensure efficient expression in E. coli. To improve expression efficiency, we optimized the gene sequence and sent it to a biotechnology company for synthesis. Next, the synthesized Bpul gene was inserted into the pET23b expression vector via EcoRI and XhoI double digestion. The Bpul gene was precisely positioned at the multiple cloning site (MCS) of the vector, ensuring efficient expression . After constructing the recombinant plasmid, it was transformed into E. coli BL21, and positive clones were confirmed through antibiotic selection. Ultimately, we successfully obtained the engineered strain BL21/pET23b-Bpul, which expresses the laccase.

Figure 1. The gene circuit of Bpul.
Characterization
Laccase activity assay

Figure 2. Gel image of the Bgls and Bupl. The Bgls gene is related to cellulase and will be studied in later experiments.
The experimental procedure was as follows: 100 mL of overnight culture of the engineered strain was collected, and its OD600 was measured. After centrifugation at 10,000 rpm for 1 min, the bacterial cells were resuspended in 20 mL of Britton-Robinson (BR) buffer and then sonicated to disrupt the cells. The supernatant was collected after centrifuging at 10,000 rpm for 20 min at 4℃, yielding the crude enzyme solution. 100 μL of the crude enzyme solution was taken, and the total protein concentration was determined using the Bradford assay, adjusting the concentration to 100 μg/mL. To assess laccase activity, 1 mM ABTS was used as the substrate, and the reaction was incubated at 37℃. The oxidation of ABTS, indicated by an increase in absorbance at 420 nm, was measured using a microplate reader. The results demonstrate that the Bpul gene from Bacillus pumilus was successfully expressed in E. coli BL21 and exhibited strong laccase activity. Compared to the control groups, the engineered strain expressing laccase significantly accelerated the degradation of ABTS, confirming its potential for degrading indigo dye in denim applications.

Figure 3. Absorbance at 420nm for different strains. As shown in the Figure 3, the absorbance values at 420 nm for the wild-type BL21, the BL21/pET23b empty vector, and the BL21/p23b-Bpul strains were compared. Both the wild-type BL21 and the BL21/pET23b strains showed negligible laccase activity, with absorbance values close to zero. In contrast, the BL21/p23b-Bpul strain exhibited a significant increase in absorbance, reaching 2.17, indicating that the expressed laccase effectively catalyzed the oxidation of ABTS.
Laccase catalytic time assay
First, 100 μL of crude enzyme solution was collected, and the total protein concentration was measured using the Bradford protein assay. The protein concentration was adjusted to 100 μg/mL using BR buffer. Then, 1 mM ABTS was added to 200 μL of the enzyme solution. The reaction mixture was incubated at 37℃ in a sealed environment, and the absorbance at 420 nm was measured at different time points: 5 min, 10 min, 15 min, 20 min, 25 min, and 30 min, using a microplate reader.The optimal catalytic time forlaccase is 15 min. This finding is valuable for applications in denim treatment, as it helps optimize the enzyme’s use in degrading indigo dye efficiently within a controlled timeframe.

Figure 4. Absorbance at 420 nm for ABTS degradation by laccase over time. As shown in Figure 4, the absorbance values at 420 nm, reflecting the degradation of ABTS by laccase, varied across the different time intervals. The control group (wild-type E. coli BL21) exhibited absorbance values close to zero throughout the experiment. In contrast, the E. coli BL21-Bpul strain expressing laccase showed a rapid increase in absorbance during the early stages of the reaction. The absorbance reached its peak at approximately 2.05 at 15 min. After 15 min, the absorbance remained stable.
Influence of temperature on laccase activity
100 μL of previously prepared crude enzyme solution was used. The protein concentration was measured using the Bradford protein assay and adjusted to 100 μg/mL. A 100 mM ABTS solution was prepared with distilled water, and 1 mM ABTS was added to 200 μL of the enzyme solution. Laccase activity was tested at different temperatures (25℃, 37℃, 45℃, 55℃, 70℃) using a thermostat water bath. The absorbance at 420 nm was measured over a 30-min period to assess ABTS degradation.The results showed that the optimal temperature for laccase activity is 55℃, where the enzymatic reaction efficiency is highest.

Figure 5. Laccase activity at different temperatures. The results showed that laccase activity increased with rising temperature and reached its peak at 55℃, with an absorbance of 2.93. Beyond 55℃, the activity dropped sharply, with absorbance falling to 1.65 at 70℃.
Influence of pH on laccase activity
The engineered bacteria were resuspended in BR buffer with varying pH values from 4 to 9. After disrupting the cells, the supernatant containing the crude enzyme was collected by centrifugation at 10,000 rpm for 20 min at 4℃. Then, 1 mM ABTS was added to the enzyme solution, and the reactions were incubated at 37℃ for 30 min in a sealed environment. Absorbance at 420 nm was measured using a microplate reader to assess laccase activity.The experimental results indicate that the optimal pH for laccase activity is pH=5, where the enzyme exhibits the highest catalytic efficiency.

Figure 6. Laccase activity at different pH. As shown in Figure 6, the activity of laccase varied across the pH range tested. The enzyme's activity increased as the pH rose from 4 to 5, reaching its peak at pH=5, with absorbance value of approximately 2.65. Beyond pH=5, the activity decreased rapidly, indicating that the enzyme becomes less efficient as the pH increases toward neutral and alkaline conditions.
Influence of copper irons on laccase activity
This experiment aims to study the effect of copper ions on laccase activity, as copper ions serve as the active center of laccase. By determining the optimal copper ion concentration, we can assess whether adding copper chloride to the final product can enhance the enzyme's catalytic efficiency. A 100 mM CuCl2 solution was prepared, and different concentrations of CuCl2 (0, 0.1, 0.25, 0.5, and 1 mM) were added to the ABTS catalytic reaction system to evaluate the effect of copper ions on laccase activity. The reactions were incubated at 37℃, pH=5, for 30 min, and the absorbance at 420 nm was measured to assess enzyme activity.The results indicate that 0.5 mM is the optimal copper ion concentration to enhance laccase activity. Based on this finding, we can add an appropriate concentration of copper ions to our laccase products to improve enzymatic activity.

Figure 7. Effect of different concentrations of copper ions on laccase activity.The experimental results showed that laccase activity increased as the copper ion concentration increased from 0 mM to 0.5 mM, with the highest activity observed at 0.5 mM. However, when the concentration increased to 1 mM, laccase activity decreased.
Testing the degradation of indigo by laccase
Previous experiments demonstrated laccase activity but did not confirm the enzyme's ability to degrade indigo. Therefore, this experiment aims to test laccase's effectiveness in degrading indigo dye. To improve the solubility of indigo, 10 mM indigo (SM4168, Beyotime) was dissolved in dimethyl sulfoxide (DMSO). A 1 mM indigo solution was added to 200 μL of crude enzyme samples (BR buffer, pH=5). A standard curve was established using different concentrations of indigo (0 mM, 0.25 mM, 0.5 mM, 1 mM, 1.5 mM, and 3 mM). The experiment was performed in a 96-well plate, sealed, and incubated at 37℃ for 2 hours. Each group was done in triplicate, and the optical density was measured at 620 nm, with indigo concentrations calculated based on the standard curve.The experimental results demonstrate that the BL21/Bpul strain, which expresses the laccase gene, significantly degrades indigo more effectively than the BL21 control strain. This confirms that laccase has strong catalytic activity in indigo degradation.

Figure 8. Effect of laccase on indigo degradation. As shown in Figure 8, after 2 hours of incubation, the indigo concentration in the BL21 strain averaged 0.95 mM, while in the BL21/Bpul strain it averaged 0.51 mM. The indigo concentration in the BL21 strain was significantly higher than that in the BL21/Bpul strain, indicating that the BL21/Bpul strain has a stronger ability to degrade indigo.
Potential Application Directions
This experiment demonstrated that the Bpul gene from Bacillus pumilus exhibits the highest laccase activity under the following conditions: temperature of 55°C, pH=5, copper ion concentration of 0.5 mM, and substrate ABTS concentration of 0.5 mM, with an optimal reaction time of 15 minutes. It effectively degrades indigo dye. In the future, the crude enzyme solution containing laccase could be processed into dry powder for industrial production, addressing the drawbacks of current jeans whitening technologies, such as safety concerns, environmental harm, immaturity of methods, and high costs.
This new product would eliminate the need for harmful chemical bleaching agents, offering an eco-friendly and gentle treatment method. Additionally, Bpul laccase dry powder is easy to transport and store, poses no threat to the environment or human health, and could provide consumers with greater freedom to DIY their own jeans. It allows for more precise control over the bleaching process, reduces fabric damage, and improves both the sustainability and quality of jeans production, making it a promising development for the future.
References
Mo Y, Lao H I, Au S W, et al. Expression, secretion and functional characterization of three laccases in E. coli[J]. Synthetic and Systems Biotechnology, 2022, 7(1): 474-480.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 123
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 490
Illegal NgoMIV site found at 958
Illegal AgeI site found at 246
Illegal AgeI site found at 1123 - 1000COMPATIBLE WITH RFC[1000]