Difference between revisions of "Part:BBa K3147004"
(→I : parts BBa_K3147004 function) |
|||
| (2 intermediate revisions by 2 users not shown) | |||
| Line 5: | Line 5: | ||
| − | ===I : parts BBa_K3147004 | + | ===I : parts BBa_K3147004 function=== |
| − | The Montpellier 2019 team made this reporter gene construct in order to obtain a positive control for TEV mediated proteolysis of the ssrA tag fused the mRFP1 reporter | + | The Montpellier 2019 team made this reporter gene construct in order to obtain a positive control for TEV mediated proteolysis of the ssrA tag fused the mRFP1 reporter [[Part:BBa_ K3147000]]. This construction produces mRFP1 fused in C-ter to a sequence corresponding to a TEV cutting site after cleavage (ENLYFQ). |
<div align="center">[[File:design2K3147004.png|650px]]</div> | <div align="center">[[File:design2K3147004.png|650px]]</div> | ||
| Line 19: | Line 19: | ||
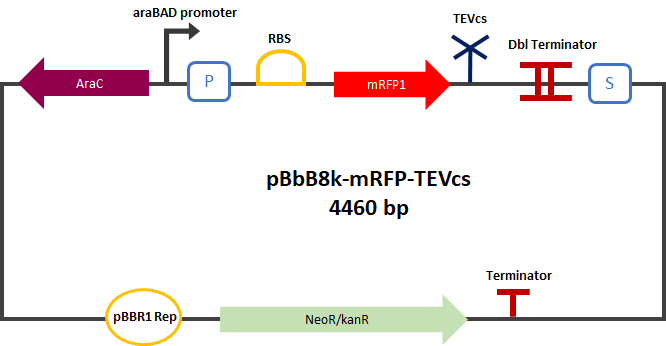
<div align="center">[[File:PlasmideK3147004.png|400px]]</div> | <div align="center">[[File:PlasmideK3147004.png|400px]]</div> | ||
| − | <div align="center"><b>Figure 2</b>: mRFP1-TEVcs | + | <div align="center"><b>Figure 2</b>: mRFP1-TEVcs reporter gene in its pBbB8k-GFP backbone.</div> |
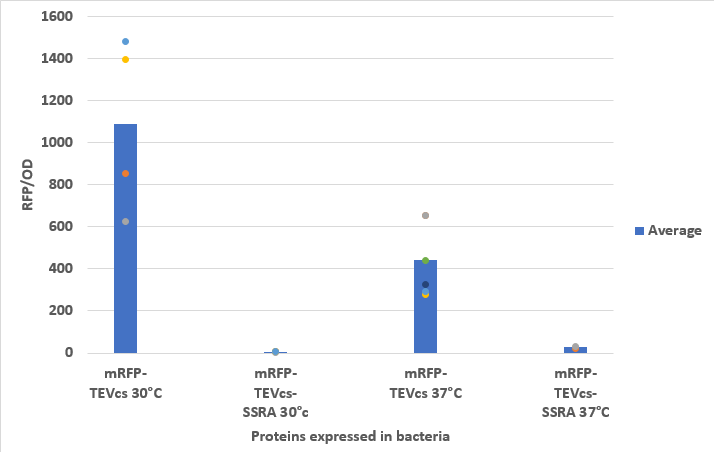
| − | We compared the basal fluorescence of the strain NEB10β of E. coli transformed with the mRFP1-TEVcs construction to an E. coli NEB10β strain transformed with the mRFP1-TEVcs-ssrA construction. Fluorescence was quantified after arabinose induction at 1% with a plate reader overnight. Here are the fluorescence of mRFP-TEVcs- | + | We compared the basal fluorescence of the strain NEB10β of E. coli transformed with the mRFP1-TEVcs construction to an E. coli NEB10β strain transformed with the mRFP1-TEVcs-ssrA construction. Fluorescence was quantified after arabinose induction at 1% with a plate reader overnight. Here are the fluorescence of mRFP-TEVcs-ssrA and mRFP-TEVcs at 30°C and 37°C. |
<div align="center">[[File:resultK3147003.png|400px]]</div> | <div align="center">[[File:resultK3147003.png|400px]]</div> | ||
| − | <div align="center"><b>Figure 3</b> :Measurement of the fluorescence at 30°C and 37°C of bacteria expressing mRFP-TEVcs or mRFP-TEVcs- | + | <div align="center"><b>Figure 3</b> :Measurement of the fluorescence at 30°C and 37°C of bacteria expressing mRFP-TEVcs or mRFP-TEVcs-ssrA</div> |
==Reference== | ==Reference== | ||
Latest revision as of 16:29, 20 October 2019
I : parts BBa_K3147004 function
The Montpellier 2019 team made this reporter gene construct in order to obtain a positive control for TEV mediated proteolysis of the ssrA tag fused the mRFP1 reporter Part:BBa_ K3147000. This construction produces mRFP1 fused in C-ter to a sequence corresponding to a TEV cutting site after cleavage (ENLYFQ).
II. Proof of function
This construction was cloned by Gibson Assembly in a pBbB8k (https://www.addgene.org/35363) backbone under the control of a pBAD promoter.
We compared the basal fluorescence of the strain NEB10β of E. coli transformed with the mRFP1-TEVcs construction to an E. coli NEB10β strain transformed with the mRFP1-TEVcs-ssrA construction. Fluorescence was quantified after arabinose induction at 1% with a plate reader overnight. Here are the fluorescence of mRFP-TEVcs-ssrA and mRFP-TEVcs at 30°C and 37°C.
Reference
[1] Levraud, Jean-Pierre et al. 2007. « Identification of the Zebrafish IFN Receptor: Implications for the Origin of the Vertebrate IFN System ». The Journal of Immunology 178(7): 4385‑94. doi:10.4049/jimmunol.178.7.4385
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 691
- 1000COMPATIBLE WITH RFC[1000]