Part:BBa_K3147000
I : parts BBa_K3147000 function
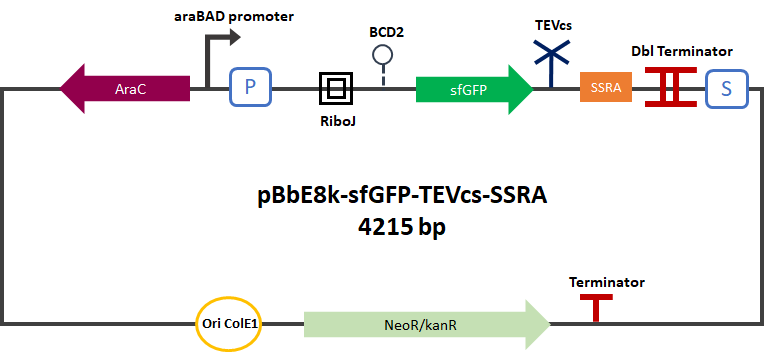
The Montpellier 2019 team submitted a reporter gene construction in order to carry out their proof of concept. This construction produces an sfGFP(bs) [1] [2] [3] Part:BBa_K1365020 fused in C-terminal with a fast degradation tag called ssrA [4] [5] Part:BBa_M0050. The TEV cutting site Part:BBa_J18918 has been added between the sfGFP and the ssrA tag. This construction can be used as a reporter for proteolysis activity by TEV. In the presence of TEV the ssrA is cleaved and sfGFP is not degraded anymore.
II. Proof of function
As a positive control, we built a construct simulating a ssrA cleavage by the TEV, by expressing sfGFP with a C-terminal sequence corresponding to the cleaved TEV recognition Part:BBa_K3147001. We expressed this part in a plasmid under the control of the arabinose promoter: pBbE8K-RFP backbone (https://www.addgene.org/35327/). We cloned it by Gibson Assembly.
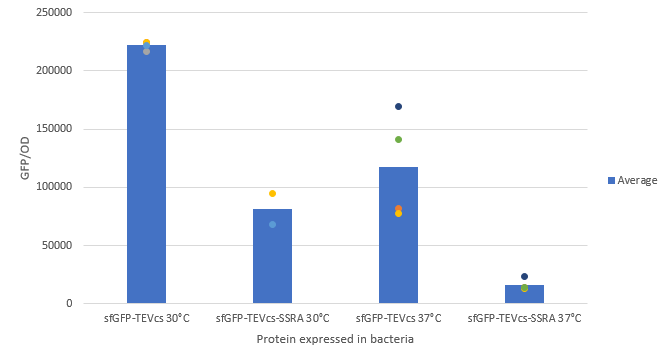
We compared the basal fluorescence of the E. coli strain NEB10β transformed with the sfGFP-TEVcs construction and the E. coli NEB10β transformed with the sfGFP-TEVcs-ssrA construction. Fluorescence was measured on a plate reader after overnight induction with 1% arabinose.
Below are the fluorescence measurements of the sfGFP-TEVcs-ssrA and of the sfGFP-TEVcs at 30 and 37°C. We can see that the ssrA tag is causing a lot of degradation of the protein. We can see that the ssrA system is more efficient at 37C as described in part M0050 characterization.
Reference:
[1] McGinness, Baker, Sauer. 2006. Mol. Cell. 22:701.
[2] Overkamp, W. et al. (2013) Benchmarking various green fluorescent protein variants in Bacillus subtilis, Streptococcus pneumoniae, and Lactococcus lactis for live cell imaging. Appl. About. Microbiol. 79: 6481-6490
[3] Sarah Guiziou et al. 2016. “A part toolbox to tune genetic expression in Bacillus subtilis” Nucleic Acids Research, 2016, Vol. 44, No. 15 7495–7508.
[4] Fernandez-Rodriguez, Jesus, et Christopher A. Voigt. 2016. « Post-TranslationalControl of Genetic Circuits Using Potyvirus Proteases ». Nucleic Acids Research 44(13): 6493‑6502.
[5] Sunohara, T., Abo, T., Inada, T., & Aiba, H. (2002). The C-terminal amino acid sequence of nascent peptide is a major determinant of SsrA tagging at all three stop codons. RNA (New York, N.Y.), 8(11), 1416–1427. doi:10.1017/s1355838202020198
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 41
Illegal NheI site found at 53
Illegal NheI site found at 76 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 82
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |