Difference between revisions of "Part:BBa K2997007"
| (10 intermediate revisions by 4 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2997007 short</partinfo> | <partinfo>BBa_K2997007 short</partinfo> | ||
| − | In the human gut microbiome, <i>Clostridium</i> related species have been reported to have the highest conversion capability of fermenting tyrosine into p- | + | In the human gut microbiome, <i>Clostridium</i> related species have been reported to have the highest conversion capability of fermenting tyrosine into <i>p</i>-Cresol<sup>[1]</sup>. To target the root of <i>p</i>-Cresol accumulation, reducing the population of <i>Clostridium</i> is needed. We decided to use <i>C. difficile</i> as a model of <i>p</i>-Cresol producing bacteria because it is a popular research target due to its notorious infectious ability. |
| − | Luckily, iGEM NCKU 2019 was kindly supported by one of our PIs, Professor | + | Luckily, iGEM NCKU-Tainan 2019 was kindly supported by one of our PIs, Professor Huang, an assistant professor from the Microbiology and Immunology Department, who is currently devoting himself to the field of developing a novel therapeutic approach for <i>C. difficile</i> infection. He kindly provided us with a plasmid containing a bacteriocin gene (CBM-B) that was proven in his lab to have bactericidal activity against certain strains of <i>Clostridium</i>, including <i>C. difficile</i>. |
| − | As proof of concept that bacteriocin is able to inhibit <i>Clostridium</i> growth, we did a spot-on-lawn assay using purified bacteriocin protein provided by advisors to observe the inhibition zone formation. As shown in Fig.1 below, a clear inhibition zone formed in the middle of the BHI plate streak with <i>C. difficile</i> R20291 strain. | + | As proof of the concept that bacteriocin is able to inhibit <i>Clostridium</i> growth, we did a spot-on-lawn assay using purified bacteriocin protein provided by advisors to observe the inhibition zone formation. As shown in Fig.1 below, a clear inhibition zone formed in the middle of the BHI plate streak with <i>C. difficile</i> R20291 strain. |
<html> | <html> | ||
| Line 19: | Line 19: | ||
| − | To further incorporate bacteriocin into our project, we wanted to integrate the bacteriocin into a live therapeutic drug instead of having patients consuming purified bacteriocins. Thus, we decided to fuse our CBM-B with a secretion tag, YebF. YebF is a secretory protein with unknown function in <i>E. coli</i>[ | + | To further incorporate bacteriocin into our project, we wanted to integrate the bacteriocin into a live therapeutic drug instead of having patients consuming purified bacteriocins. Thus, we decided to fuse our CBM-B with a secretion tag, YebF. YebF is a secretory protein with unknown function in <i>E. coli</i><sup>[2]</sup>. We first amplified the <i>yebF</i> fragments from the genome of <i>E. coli</i> MG1655 and the CBM-B fragment from the plasmid kindly provided by Professor Huang. Literature has reported that the CBM-B gene encodes a bacteriocin protein that will perform self-cleavage, and only the C-terminus fragment has bactericidal properties. So, we designed two versions of CBM-B construct by separately amplifying the full version of CBM-B gene and also the s(short)CBMB that encodes the C-terminus protein region only. Concerned that the fusion of sCBM-B with YebF may affect its bactericidal function, two linker sequences were introduced between <i>yebF</i> and sCBM-B using overhang primers. We also used PCR to amplify the [[Part:BBa_K880005 | BBa_K880005]] promoter from iGEM distribution kit, and used overextension PCR to piece these fragments together, before ligating them into pSB3K3 plasmid. |
<html> | <html> | ||
<br> | <br> | ||
| Line 28: | Line 28: | ||
</html> | </html> | ||
| − | Fig. 2. (a) Schematics of PCR strategy for each reaction. (b) 1.5% Agarose Gel figure show PCR result. M: Marker; Lane 1: Promoter | + | Fig. 2. (a) Schematics of PCR strategy for each reaction. (b) 1.5% Agarose Gel figure show PCR result. M: Marker; Lane 1: Promoter and RBS; Lane 2: Promoter and RBS; Lane 3: CBM-B; Lane 4: GS linker - sCBM-B; Lane 5: TB linker - sCBM-B; Lane 6: <i>yebF</i> - GS linker overhang; Lane 7: <i>yebF</i> - TB linker overhang; Lane 8:<i>yebF</i> - CBM-B overhang. |
<html> | <html> | ||
| Line 39: | Line 39: | ||
Fig. 3. (a) Schematic of PCR strategy for each reaction. (b) 1.5 % Agarose Gel figure show PCR result. M: Marker; Lane 1: PCR product of BBa_K2997000; Lane 2: PCR product of BBa_K2997005; Lane 3: PCR product of BBa_K2997006; Lane 4: PCR product of BBa_K2997007 | Fig. 3. (a) Schematic of PCR strategy for each reaction. (b) 1.5 % Agarose Gel figure show PCR result. M: Marker; Lane 1: PCR product of BBa_K2997000; Lane 2: PCR product of BBa_K2997005; Lane 3: PCR product of BBa_K2997006; Lane 4: PCR product of BBa_K2997007 | ||
| − | However, after several attempts of constructing the yebF-bacteriocin plasmid, mutations kept appearing in the coding region. We will further complete yebF-bacteriocin construct in the future. We | + | However, after several attempts of constructing the <i>yebF</i> - bacteriocin plasmid, mutations kept appearing in the coding region. We will further complete <i>yebF</i> - bacteriocin construct in the future. We hypothesized that adding the <i>yebF</i> secretion tag into the construct is toxic to the bacteria. |
<html> | <html> | ||
| Line 48: | Line 48: | ||
<br> | <br> | ||
</html> | </html> | ||
| − | Fig. 4. Alignment results showing 1bp deletion causing frameshifting in the coding region of yebF- | + | Fig. 4. Alignment results showing 1bp deletion causing frameshifting in the coding region of <i>yebF</i> - GS linker - sCBM-B construct. Red triangular indicates the deletion base pair. |
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 63: | Line 64: | ||
<!-- --> | <!-- --> | ||
===Reference=== | ===Reference=== | ||
| − | [1] Saito, Y., Sato, T., Nomoto, K., & Tsuji, H. (2018). Identification of phenol- and p- | + | [1] Saito, Y., Sato, T., Nomoto, K., & Tsuji, H. (2018). Identification of phenol- and <i>p</i>-Cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiology Ecology, 94(9). |
| − | [2] | + | [2] Zhang, G., Brokx, S., & Weiner, J. H. (2005). Extracellular accumulation of recombinant proteins fused to the carrier protein YebF in Escherichia coli. Nature Biotechnology, 24(1), 100–104. |
Latest revision as of 11:12, 13 December 2019
K880005 - yebF - CBM-B
In the human gut microbiome, Clostridium related species have been reported to have the highest conversion capability of fermenting tyrosine into p-Cresol[1]. To target the root of p-Cresol accumulation, reducing the population of Clostridium is needed. We decided to use C. difficile as a model of p-Cresol producing bacteria because it is a popular research target due to its notorious infectious ability.
Luckily, iGEM NCKU-Tainan 2019 was kindly supported by one of our PIs, Professor Huang, an assistant professor from the Microbiology and Immunology Department, who is currently devoting himself to the field of developing a novel therapeutic approach for C. difficile infection. He kindly provided us with a plasmid containing a bacteriocin gene (CBM-B) that was proven in his lab to have bactericidal activity against certain strains of Clostridium, including C. difficile.
As proof of the concept that bacteriocin is able to inhibit Clostridium growth, we did a spot-on-lawn assay using purified bacteriocin protein provided by advisors to observe the inhibition zone formation. As shown in Fig.1 below, a clear inhibition zone formed in the middle of the BHI plate streak with C. difficile R20291 strain.
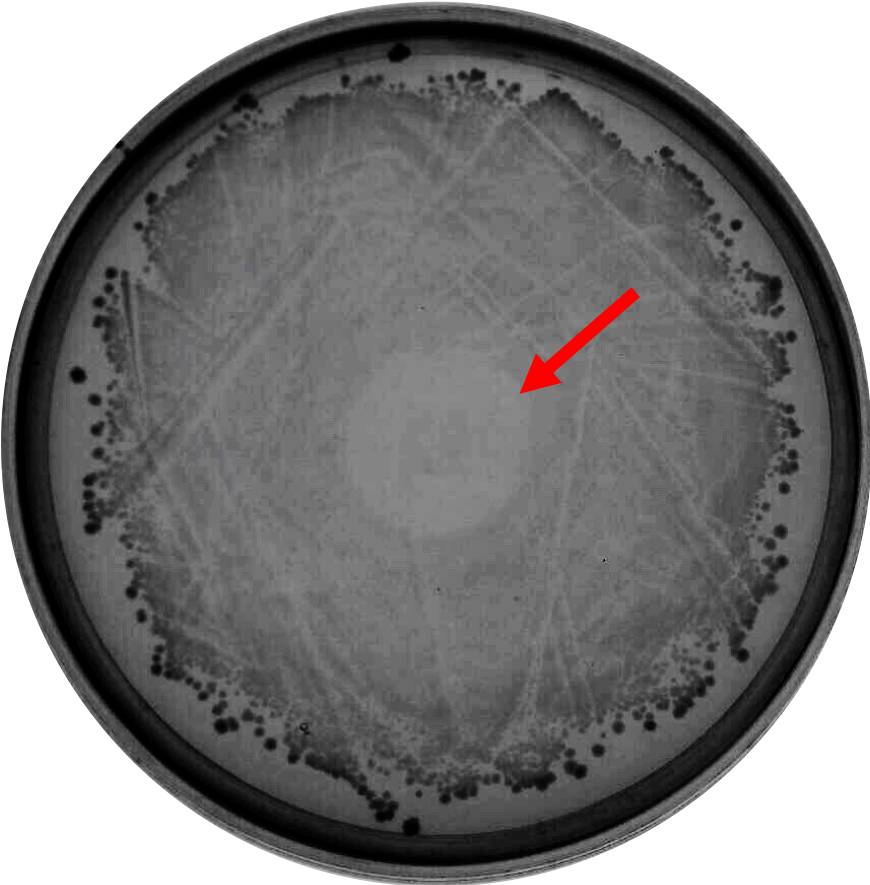
Fig. 1. Spot-on-lawn test using 5 μl purified bacteriocin, inhibition zone formation in the middle of the plate can clearly be seen.
To further incorporate bacteriocin into our project, we wanted to integrate the bacteriocin into a live therapeutic drug instead of having patients consuming purified bacteriocins. Thus, we decided to fuse our CBM-B with a secretion tag, YebF. YebF is a secretory protein with unknown function in E. coli[2]. We first amplified the yebF fragments from the genome of E. coli MG1655 and the CBM-B fragment from the plasmid kindly provided by Professor Huang. Literature has reported that the CBM-B gene encodes a bacteriocin protein that will perform self-cleavage, and only the C-terminus fragment has bactericidal properties. So, we designed two versions of CBM-B construct by separately amplifying the full version of CBM-B gene and also the s(short)CBMB that encodes the C-terminus protein region only. Concerned that the fusion of sCBM-B with YebF may affect its bactericidal function, two linker sequences were introduced between yebF and sCBM-B using overhang primers. We also used PCR to amplify the BBa_K880005 promoter from iGEM distribution kit, and used overextension PCR to piece these fragments together, before ligating them into pSB3K3 plasmid.
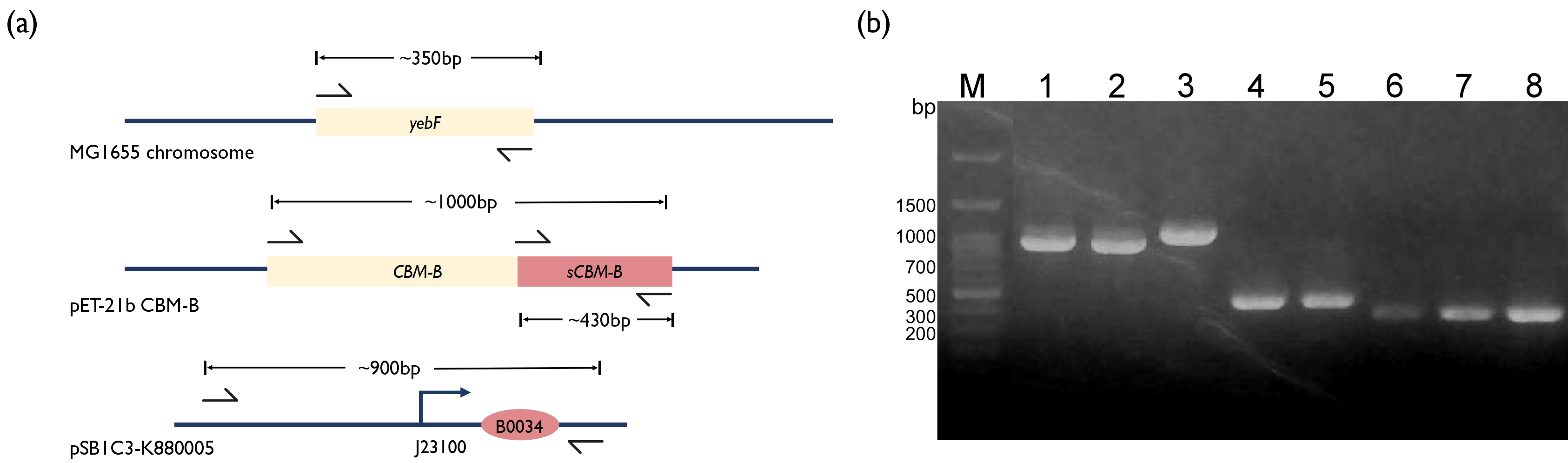
Fig. 2. (a) Schematics of PCR strategy for each reaction. (b) 1.5% Agarose Gel figure show PCR result. M: Marker; Lane 1: Promoter and RBS; Lane 2: Promoter and RBS; Lane 3: CBM-B; Lane 4: GS linker - sCBM-B; Lane 5: TB linker - sCBM-B; Lane 6: yebF - GS linker overhang; Lane 7: yebF - TB linker overhang; Lane 8:yebF - CBM-B overhang.
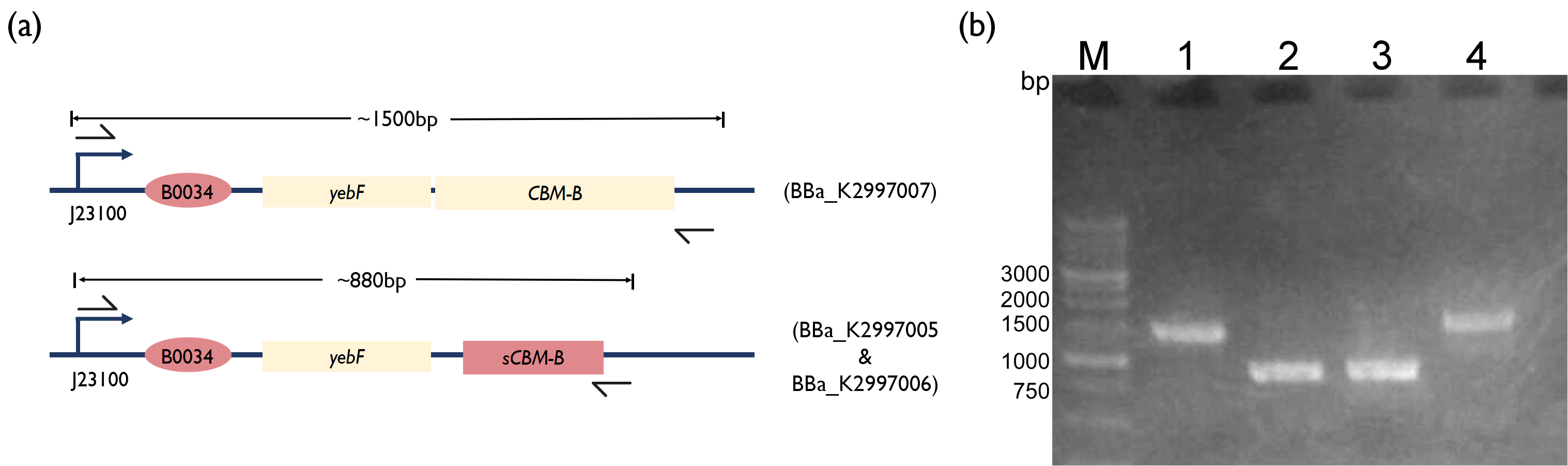
Fig. 3. (a) Schematic of PCR strategy for each reaction. (b) 1.5 % Agarose Gel figure show PCR result. M: Marker; Lane 1: PCR product of BBa_K2997000; Lane 2: PCR product of BBa_K2997005; Lane 3: PCR product of BBa_K2997006; Lane 4: PCR product of BBa_K2997007
However, after several attempts of constructing the yebF - bacteriocin plasmid, mutations kept appearing in the coding region. We will further complete yebF - bacteriocin construct in the future. We hypothesized that adding the yebF secretion tag into the construct is toxic to the bacteria.

Fig. 4. Alignment results showing 1bp deletion causing frameshifting in the coding region of yebF - GS linker - sCBM-B construct. Red triangular indicates the deletion base pair.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal SpeI site found at 819
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30
Illegal SpeI site found at 819 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 169
- 23INCOMPATIBLE WITH RFC[23]Illegal SpeI site found at 819
- 25INCOMPATIBLE WITH RFC[25]Illegal SpeI site found at 819
- 1000COMPATIBLE WITH RFC[1000]
Reference
[1] Saito, Y., Sato, T., Nomoto, K., & Tsuji, H. (2018). Identification of phenol- and p-Cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiology Ecology, 94(9).
[2] Zhang, G., Brokx, S., & Weiner, J. H. (2005). Extracellular accumulation of recombinant proteins fused to the carrier protein YebF in Escherichia coli. Nature Biotechnology, 24(1), 100–104.
