Difference between revisions of "Part:BBa K3037005"
(→4) Measurement of fluorescence) |
|||
| (90 intermediate revisions by 4 users not shown) | |||
| Line 10: | Line 10: | ||
|- | |- | ||
|'''RFC standard''' | |'''RFC standard''' | ||
| − | | | + | |[[Help:Assembly_standard_25|Freiburg RFC25 standard]] |
|- | |- | ||
|'''Backbone''' | |'''Backbone''' | ||
| − | |pSB1C3<br> | + | |[https://parts.igem.org/Help:2019_DNA_Distribution pSB1C3<br>] |
| + | |- | ||
| + | |'''Experimental Backbone''' | ||
| + | |[https://parts.igem.org/Part:BBa_K3037000 pOCC97<br>] | ||
|- | |- | ||
|'''Submitted by''' | |'''Submitted by''' | ||
| − | |Team: | + | |Team: [https://2019.igem.org/Team:TU_Dresden TU_Dresden 2019] |
|} | |} | ||
| + | |||
| Line 23: | Line 27: | ||
== Overview == | == Overview == | ||
| − | This BioBrick was designed as a composite BioBrick by the team TU Dresden 2019. It is a fusion of dCas9 [https://parts.igem.org/Part:BBa_K3037002 (BBa_K3037002)] and eGFP [https://parts.igem.org/Part:BBa_K3037006 (BBa_K3037006)]. | + | This BioBrick was designed as a composite BioBrick by the team TU Dresden 2019. It is a fusion of dCas9 [https://parts.igem.org/Part:BBa_K3037002 (BBa_K3037002)] and eGFP [https://parts.igem.org/Part:BBa_K3037006 (BBa_K3037006)][https://2019.igem.org/Team:TU_Dresden/Parts (more information).] |
| − | dCas9 can bind to any sequence of DNA | + | The dCas9 can bind to any sequence of DNA specified by its guideRNA. The eGFP (enhanced reporter protein) is used as reporter, resulting in fluorescence emission at 530 nm [1]. |
| − | Additionally the BioBrick is designed to have | + | <div style="text-align:justify;"> |
| + | Additionally, the BioBrick is designed to have a N-terminal Maltose-binding-protein (MBP) fused to it [https://parts.igem.org/Part:BBa_K3037001 (BBa_K3037001).]This improves the solubility and expression and thus, the total cytoplasmic yield of the protein. The MBP can further be used to purify the fusion protein via amylose resin and afterwards, it can be cleaved off by digestion with a PreScission protease. | ||
| + | |||
| + | The design of the here presented BioBrick is based on [https://parts.igem.org/Part:BBa_K1994025 BBa_K1994025.] The original composite BioBrick consists of a transcriptional fusion of GFP and dCas9, both having their own promoter and RBS. A terminator separates both units. However, from our perspective having two transcriptional units as a composite part makes little sense to couple dCas9 activity with an easy readout - such as GFP. Therefore, our improvement was to fuse the dCas9 with eGFP to generate a translational fusion protein. Additionally, having the MBP fused on the N-terminus facilitates purification and easy cleaving off, as already explained before. We strongly believe that the combination of these improvements will be tremendously helpful for future iGEM teams working on dCas9 based projects. Down below, our characterization of the new improved composite part. | ||
== Characterization == | == Characterization == | ||
| Line 34: | Line 41: | ||
We performed the following characterization experiments: | We performed the following characterization experiments: | ||
| − | 1) | + | <b>1)</b> Expression using pOCC97 [https://parts.igem.org/Part:BBa_K3037000 (BBa_K3037000)] in <i>E. coli</i> pRARE T7 - Proof of the correct functioning of GFP |
| − | 2) | + | <b>2)</b> Construct purification and tag-removal - Proof of the correct functioning of MBP |
| − | 3) | + | <b>3)</b> Proof of DNA-binding ability of dCas9 via Electrophoretic Mobility Shift Assay (EMSA) - Proof of the correct functioning of dCas9 |
| − | 4) | + | <b>4)</b> Measurement of fluorescence |
=== Experiments in Detail === | === Experiments in Detail === | ||
| − | ==== 1) | + | ==== 1) Expression using pOCC97 (BBa_K3037000) in <i>E. coli</i> pRARE T7 - Proof of the correct functioning of GFP ==== |
| + | As a quick and easy proof of principle experiment, we used our over-expression plasmid pOCC97 ([https://parts.igem.org/Part:BBa_K3037000 BBa_K3037000) and introduced our construct (BBa_K3037005). After inducing expression in growing <i>E.coli</i> cells with IPTG, we took a sample, transferred it into an Eppendorf tupe and showed that these <i>E.coli</i> cells glow upon excitation with UV light (Figure 1). Thus, we concluded that our construct of MBP+eGFP+dCas9 was expressed successfully. | ||
| + | [[File:T--TU_Dresden--Expressing_GFP_BBa_K3037000.png|center|200px|thumb|none|Figure 1: Expression of BBa_K3037005 in pOCC97]] | ||
| − | + | ==== 2) Construct purification and tag-removal - Proof of the correct functioning of MBP ==== | |
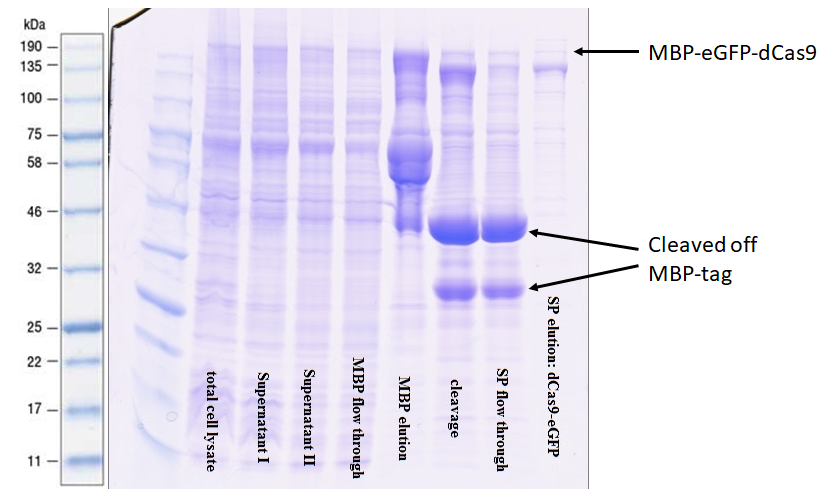
| − | + | The protein was purified via an amylose resin column with the a N-terminal-MBP-tag [https://parts.igem.org/Part:BBa_K3037001 BBa_K3037001] (Figure 2). | |
| − | |||
| − | + | [[File:T--TU_Dresden--Amilose_Resin_2_BBa_K3037005.png|center|400px|thumb|none|Figure 2: Amilose resin purification using MBP-tag. The upper arrow shows the location in the gel of this BioBrick (BBa_K3037005). The lower ones show the MBP-tag after beeing cleaved off]] | |
| − | + | The comparison of lane 4 and 5 illustrates nicely the performance of the MPP-tag with the amylose resin. Upon elution in lane 5 many truncated versions appear. This was expected since it often occurs when expressing large recombinant proteins. The high intensity of the bands shows that previously these proteins were bound to the resin as they were not in lane 4. After the digestion with 3C protease, a very strong signal appears at 42 kDa indicating that the preScission sites are intact and were recognized. The purification of the complete transcript from the cleaved off tag was achieved by cation exchange chromatography on a HiTrap SP column. | |
| − | + | ==== 3) Prove of DNA-binding ability of dCas9 via EMSA - Proof of the correct functioning of dCas9==== | |
| − | + | <b>1. Materials:</b> | |
| − | + | *100 ng of PCR amplified <span style="font-style: italic;">sry</span> gene | |
| + | *200 ng of dCas9-GFP | ||
| + | *200 ng of guide RNA specifically targeting the amplified <span style="font-style: italic;">sry</span> gene | ||
| + | *1 x Reaction buffer - 20 mM Hepes buffer (pH 7.2) | ||
| + | ** 100 mM NaCl | ||
| + | ** 5 mM MgCl2 | ||
| + | ** 0.1 mM EDTA | ||
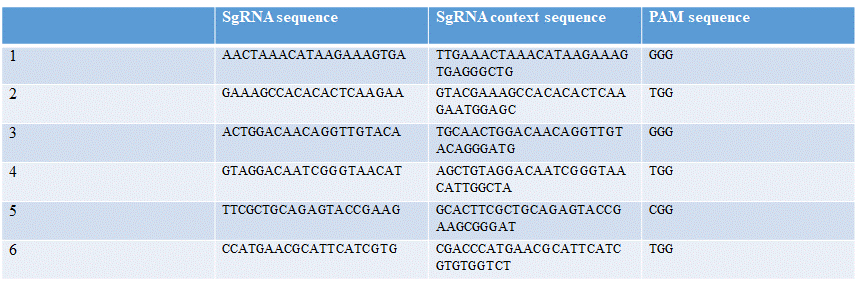
| − | Six different guide RNAs were designed for targeting different regions of <span style="font-style: italic;"> | + | Six different guide RNAs were designed for targeting different regions of <span style="font-style: italic;">sry</span> gene. Using the online tool Benchling and Fasta sequence of <span style="font-style: italic;">sry</span> gene (Table 1). |
1: AACTAAACATAAGAAAGTGA | 1: AACTAAACATAAGAAAGTGA | ||
| Line 78: | Line 92: | ||
6: CCATGAACGCATTCATCGTG | 6: CCATGAACGCATTCATCGTG | ||
| − | [[File:T--TU_Dresden--EMSA_Primers_BBa_K3037005.png| | + | [[File:T--TU_Dresden--EMSA_Primers_BBa_K3037005.png|center|400px|thumb|left|Table 1: Overview of different guide RNAs with the context of the sequence and the PAM sequence]] |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<b>2. Methods:</b> | <b>2. Methods:</b> | ||
| − | 1. We wanted to check if the overall efficiency of mobility shift increases when combinations of guide RNAs are | + | 1. We wanted to check if the overall efficiency of mobility shift increases when different combinations of guide RNAs are used. |
| − | 2. Guide RNA, dCas9-GFP and <span style="font-style: italic;"> | + | 2. Guide RNA, dCas9-GFP and <span style="font-style: italic;">sry</span> gene were incubated in reaction buffer (respective amounts mentioned in the materials section) for 37 °C for 1 hour. |
| − | 3. Post incubation, they were mixed with loading dye without SDS, 20 % glycerol in Orange G dye and loaded onto 4-20 % gradient acrylamide- TBE precast gel. | + | 3. Post incubation, they were mixed with loading dye without SDS, 20 % glycerol in Orange G dye and loaded onto a 4-20 % gradient acrylamide- TBE precast gel. Two gels were run for 2 and 3 hours at 75 V in 1 x TBE buffer. |
| − | 4. Gel was then stained using | + | 4. Gel was then stained using EtBr with 1:20000 dilution in 1x TBE for 10 minutes. |
<b>3.1 Results and Discussion of the 2 hours gel:</b> | <b>3.1 Results and Discussion of the 2 hours gel:</b> | ||
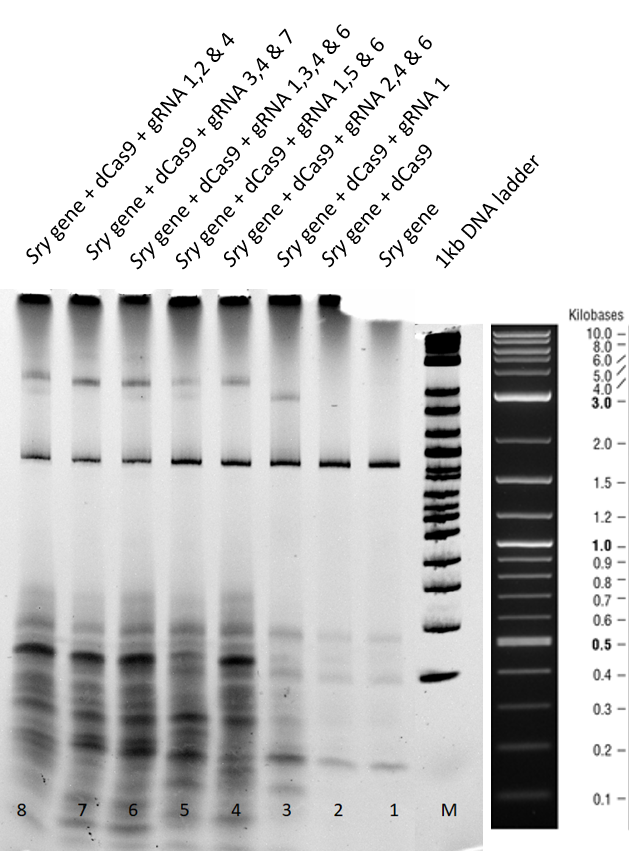
| − | [[File:T--TU_Dresden--EMSA1_BBa_K3037005.png| | + | [[File:T--TU_Dresden--EMSA1_BBa_K3037005.png|center|400px|thumb|left|Figure 3: Electrophoretic Mobility Shift Assay (EMSA) after a two hours run using dCas9 with different guideRNAs.]] |
| − | Lane 1 - There is a clear <span style="font-style: italic;"> | + | Lane 1+2 - There is a clear <span style="font-style: italic;">sry</span> band at 800 base pairs and when the <span style="font-style: italic;">sry</span> gene is incubated only with dCas9 without guideRNA. Over all, no shift is observed. |
| − | Lane | + | Lane 3 - When guideRNA 1 was incubated with the dCas9 DNA reaction mix, we saw a shift in the mobility caused by the protein DNA interaction. The binding hinders the DNA mobility. |
| − | + | Lanes 5, 6, 7, 8 and 9 – Different combinations of guide RNAs were used. From lane 7 and 8 we see the highest mobility shift. | |
| − | + | From the electromobility shift assay performed above, we conclude that our expressed dCas9-GFP protein is functional and is able to successfully bind to DNA with the help of appropriate guideRNAs. | |
| − | + | ||
| − | From the electromobility shift assay performed above, we conclude that our expressed dCas9-GFP protein is functional and is able to successfully bind to | + | |
<b>3.2 Results and Discussion of the 3 hours gel:</b> | <b>3.2 Results and Discussion of the 3 hours gel:</b> | ||
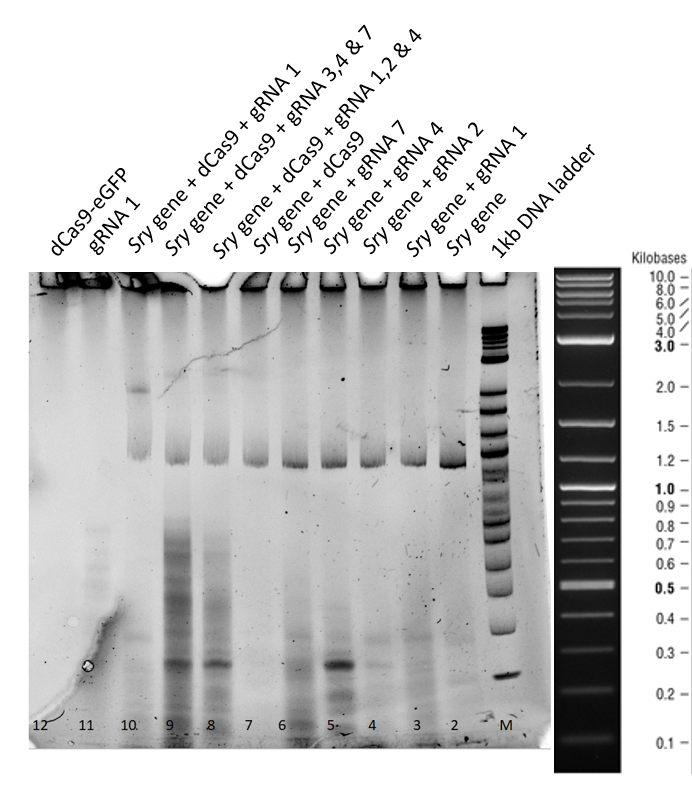
| − | [[File:T--TU_Dresden--EMSA2_BBa_K3037005.png| | + | [[File:T--TU_Dresden--EMSA2_BBa_K3037005.png|center|400px|thumb|left|Figure 4: Electrophoretic Mobility Shift Assay (EMSA) after a three hour run of the dCas9 with different guideRNAs]] |
| − | This second gel was run | + | This second gel was run longer in order to remove all the secondary structures derived from residual RNA fragments. |
| − | From lane 3 to 7, no difference in the mobility of | + | From lane 3 to 7, no difference in the mobility of <i>sry</i> gene can be seen when only guideRNA is added to the reaction mix. |
| − | In | + | In lane 8, 9 and 10 a mobility shift of the gene can be observed and in lane 11, where only guideRNA was loaded no bands were obtained. |
| − | + | ||
| − | + | ||
| + | In lane 12, dCas9 is in the stacking part of gel, corresponding to higher molecular weight. | ||
<b>4. Conclusions:</b> | <b>4. Conclusions:</b> | ||
| − | - We have a functional dCas9 expressed, which is able bind successfully to <span style="font-style: italic;"> | + | - We have a functional dCas9 expressed, which is able to bind successfully to <span style="font-style: italic;">sry</span> gene with the help of specific guideRNAs. |
| − | - dCas9 on its own is unable to bind to <span style="font-style: italic;"> | + | - dCas9 on its own is unable to bind to <span style="font-style: italic;">sry</span> gene, proving that for binding at least one appropriate guideRNA is required. |
| − | - | + | - GuideRNAs on their own are unable to cause a mobility shift of the <span style="font-style: italic;">sry</span> gene. |
| + | ==== 4) Measurement of fluorescence ==== | ||
| + | |||
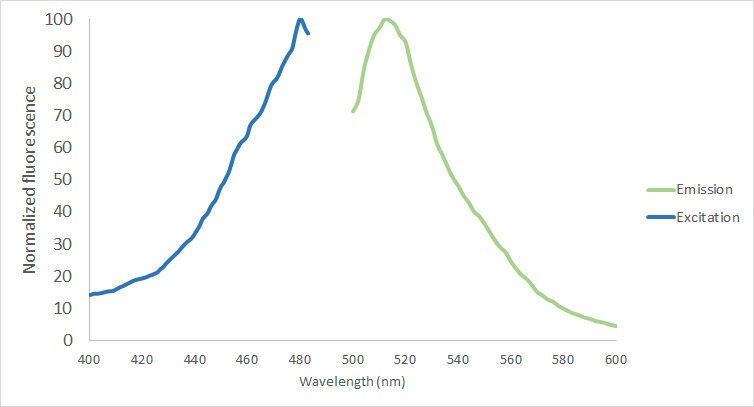
| + | The protein functionality of eGFP in the fusion protein was analyzed in the protein purified using the MBP-tag. First of all, the excitation and emission spectrum were measured with Tecan Plate Reader (Bandwith 20 nm) (Figure 5). The excitation maxima was screened from 400 to 490, being the final excitation maxima 480 nm. The emission maxima was measured from 500 to 600 nm with a final result of 512 nm. | ||
| + | |||
| + | |||
| + | [[File:T--TU_Dresden--Fluorescence_BBa_K3037005.png|center|400px|thumb|left|Figure 5: Fluorescense spectrum measurement. Excitation maxima 480 nm. Emission maxima 512 nm.]] | ||
== Sequence == | == Sequence == | ||
| − | + | NOTE: Please be aware, that by combining all the basic parts used for this composite part, the registry automatically inserted a RFC23 scar. However, the design and our cloning strategy is based on RFC25. | |
| + | <partinfo>BBa_K3037005 SequenceAndFeatures</partinfo> | ||
== References == | == References == | ||
| − | [1] | + | [1] https://www.biotek.com/resources/technical-notes/excitation-and-emission-of-green-fluorescent-proteins/ |
[2] https://www.genscript.com/bacterial-soluble-protein-expression-MBP-tag.html | [2] https://www.genscript.com/bacterial-soluble-protein-expression-MBP-tag.html | ||
[3] https://www.gelifesciences.com/en/us/shop/chromatography/resins/affinity-tagged-protein/prescission-protease-for-gst-tag-removal-p-00248 | [3] https://www.gelifesciences.com/en/us/shop/chromatography/resins/affinity-tagged-protein/prescission-protease-for-gst-tag-removal-p-00248 | ||
Latest revision as of 02:09, 22 October 2019
MBP+eGFP+dCas9
| MBP/eGFP/dCas9 | |
|---|---|
| Function | Reporter |
| Use in | Escherichia coli |
| RFC standard | Freiburg RFC25 standard |
| Backbone | pSB1C3 |
| Experimental Backbone | pOCC97 |
| Submitted by | Team: TU_Dresden 2019 |
Contents
- 1 Overview
- 2 Characterization
- 2.1 Outline
- 2.2 Experiments in Detail
- 2.2.1 1) Expression using pOCC97 (BBa_K3037000) in E. coli pRARE T7 - Proof of the correct functioning of GFP
- 2.2.2 2) Construct purification and tag-removal - Proof of the correct functioning of MBP
- 2.2.3 3) Prove of DNA-binding ability of dCas9 via EMSA - Proof of the correct functioning of dCas9
- 2.2.4 4) Measurement of fluorescence
- 3 Sequence
- 4 References
Overview
This BioBrick was designed as a composite BioBrick by the team TU Dresden 2019. It is a fusion of dCas9 (BBa_K3037002) and eGFP (BBa_K3037006)(more information).
The dCas9 can bind to any sequence of DNA specified by its guideRNA. The eGFP (enhanced reporter protein) is used as reporter, resulting in fluorescence emission at 530 nm [1].
Additionally, the BioBrick is designed to have a N-terminal Maltose-binding-protein (MBP) fused to it (BBa_K3037001).This improves the solubility and expression and thus, the total cytoplasmic yield of the protein. The MBP can further be used to purify the fusion protein via amylose resin and afterwards, it can be cleaved off by digestion with a PreScission protease.
The design of the here presented BioBrick is based on BBa_K1994025. The original composite BioBrick consists of a transcriptional fusion of GFP and dCas9, both having their own promoter and RBS. A terminator separates both units. However, from our perspective having two transcriptional units as a composite part makes little sense to couple dCas9 activity with an easy readout - such as GFP. Therefore, our improvement was to fuse the dCas9 with eGFP to generate a translational fusion protein. Additionally, having the MBP fused on the N-terminus facilitates purification and easy cleaving off, as already explained before. We strongly believe that the combination of these improvements will be tremendously helpful for future iGEM teams working on dCas9 based projects. Down below, our characterization of the new improved composite part.
Characterization
Outline
We performed the following characterization experiments:
1) Expression using pOCC97 (BBa_K3037000) in E. coli pRARE T7 - Proof of the correct functioning of GFP
2) Construct purification and tag-removal - Proof of the correct functioning of MBP
3) Proof of DNA-binding ability of dCas9 via Electrophoretic Mobility Shift Assay (EMSA) - Proof of the correct functioning of dCas9
4) Measurement of fluorescence
Experiments in Detail
1) Expression using pOCC97 (BBa_K3037000) in E. coli pRARE T7 - Proof of the correct functioning of GFP
As a quick and easy proof of principle experiment, we used our over-expression plasmid pOCC97 ([https://parts.igem.org/Part:BBa_K3037000 BBa_K3037000) and introduced our construct (BBa_K3037005). After inducing expression in growing E.coli cells with IPTG, we took a sample, transferred it into an Eppendorf tupe and showed that these E.coli cells glow upon excitation with UV light (Figure 1). Thus, we concluded that our construct of MBP+eGFP+dCas9 was expressed successfully.
2) Construct purification and tag-removal - Proof of the correct functioning of MBP
The protein was purified via an amylose resin column with the a N-terminal-MBP-tag BBa_K3037001 (Figure 2).
The comparison of lane 4 and 5 illustrates nicely the performance of the MPP-tag with the amylose resin. Upon elution in lane 5 many truncated versions appear. This was expected since it often occurs when expressing large recombinant proteins. The high intensity of the bands shows that previously these proteins were bound to the resin as they were not in lane 4. After the digestion with 3C protease, a very strong signal appears at 42 kDa indicating that the preScission sites are intact and were recognized. The purification of the complete transcript from the cleaved off tag was achieved by cation exchange chromatography on a HiTrap SP column.
3) Prove of DNA-binding ability of dCas9 via EMSA - Proof of the correct functioning of dCas9
1. Materials:
- 100 ng of PCR amplified sry gene
- 200 ng of dCas9-GFP
- 200 ng of guide RNA specifically targeting the amplified sry gene
- 1 x Reaction buffer - 20 mM Hepes buffer (pH 7.2)
- 100 mM NaCl
- 5 mM MgCl2
- 0.1 mM EDTA
Six different guide RNAs were designed for targeting different regions of sry gene. Using the online tool Benchling and Fasta sequence of sry gene (Table 1).
1: AACTAAACATAAGAAAGTGA
2: GAAAGCCACACACTCAAGAA
3: ACTGGACAACAGGTTGTACA
4: GTAGGACAATCGGGTAACAT
5: TTCGCTGCAGAGTACCGAAG
6: CCATGAACGCATTCATCGTG
2. Methods:
1. We wanted to check if the overall efficiency of mobility shift increases when different combinations of guide RNAs are used.
2. Guide RNA, dCas9-GFP and sry gene were incubated in reaction buffer (respective amounts mentioned in the materials section) for 37 °C for 1 hour.
3. Post incubation, they were mixed with loading dye without SDS, 20 % glycerol in Orange G dye and loaded onto a 4-20 % gradient acrylamide- TBE precast gel. Two gels were run for 2 and 3 hours at 75 V in 1 x TBE buffer.
4. Gel was then stained using EtBr with 1:20000 dilution in 1x TBE for 10 minutes.
3.1 Results and Discussion of the 2 hours gel:
Lane 1+2 - There is a clear sry band at 800 base pairs and when the sry gene is incubated only with dCas9 without guideRNA. Over all, no shift is observed.
Lane 3 - When guideRNA 1 was incubated with the dCas9 DNA reaction mix, we saw a shift in the mobility caused by the protein DNA interaction. The binding hinders the DNA mobility.
Lanes 5, 6, 7, 8 and 9 – Different combinations of guide RNAs were used. From lane 7 and 8 we see the highest mobility shift.
From the electromobility shift assay performed above, we conclude that our expressed dCas9-GFP protein is functional and is able to successfully bind to DNA with the help of appropriate guideRNAs.
3.2 Results and Discussion of the 3 hours gel:
This second gel was run longer in order to remove all the secondary structures derived from residual RNA fragments.
From lane 3 to 7, no difference in the mobility of sry gene can be seen when only guideRNA is added to the reaction mix.
In lane 8, 9 and 10 a mobility shift of the gene can be observed and in lane 11, where only guideRNA was loaded no bands were obtained.
In lane 12, dCas9 is in the stacking part of gel, corresponding to higher molecular weight.
4. Conclusions:
- We have a functional dCas9 expressed, which is able to bind successfully to sry gene with the help of specific guideRNAs.
- dCas9 on its own is unable to bind to sry gene, proving that for binding at least one appropriate guideRNA is required.
- GuideRNAs on their own are unable to cause a mobility shift of the sry gene.
4) Measurement of fluorescence
The protein functionality of eGFP in the fusion protein was analyzed in the protein purified using the MBP-tag. First of all, the excitation and emission spectrum were measured with Tecan Plate Reader (Bandwith 20 nm) (Figure 5). The excitation maxima was screened from 400 to 490, being the final excitation maxima 480 nm. The emission maxima was measured from 500 to 600 nm with a final result of 512 nm.
Sequence
NOTE: Please be aware, that by combining all the basic parts used for this composite part, the registry automatically inserted a RFC23 scar. However, the design and our cloning strategy is based on RFC25.
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 3061
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 381
Illegal BamHI site found at 1930
Illegal BamHI site found at 5340 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 79
References
[2] https://www.genscript.com/bacterial-soluble-protein-expression-MBP-tag.html
[3] https://www.gelifesciences.com/en/us/shop/chromatography/resins/affinity-tagged-protein/prescission-protease-for-gst-tag-removal-p-00248