Difference between revisions of "Part:BBa K864402"
(→Contribution) |
|||
| (20 intermediate revisions by 3 users not shown) | |||
| Line 8: | Line 8: | ||
[[Image:eforred_plate_small.jpg|420px]] | [[Image:eforred_plate_small.jpg|420px]] | ||
[[Image:UUChromo.jpg|500px]] | [[Image:UUChromo.jpg|500px]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===Usage and Biology=== | ||
===Contribution=== | ===Contribution=== | ||
| Line 15: | Line 21: | ||
<br> | <br> | ||
Summary: | Summary: | ||
| − | In this contribution we characterized the visual absorbance and the fluorescence of this construct. We also tested the oxygen dependency of the | + | In this contribution, we characterized the visual absorbance and the fluorescence of this construct. We also tested the oxygen dependency of the protein expression in <i>E. coli </i> BL21 (DE3) cells. The molecular weight of eforRed was also examined with an SDS-PAGE.<br> |
Documentation: | Documentation: | ||
| + | |||
<html> | <html> | ||
| Line 24: | Line 31: | ||
<li> </html>the [https://parts.igem.org/wiki/index.php?title=Part:BBa_B0034 BBa_B0034]Ribosome binding site<html></li> | <li> </html>the [https://parts.igem.org/wiki/index.php?title=Part:BBa_B0034 BBa_B0034]Ribosome binding site<html></li> | ||
</ul> | </ul> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <p> | ||
| + | <b style="font-size:120%;">Fluorescence and absorbance</b><br> | ||
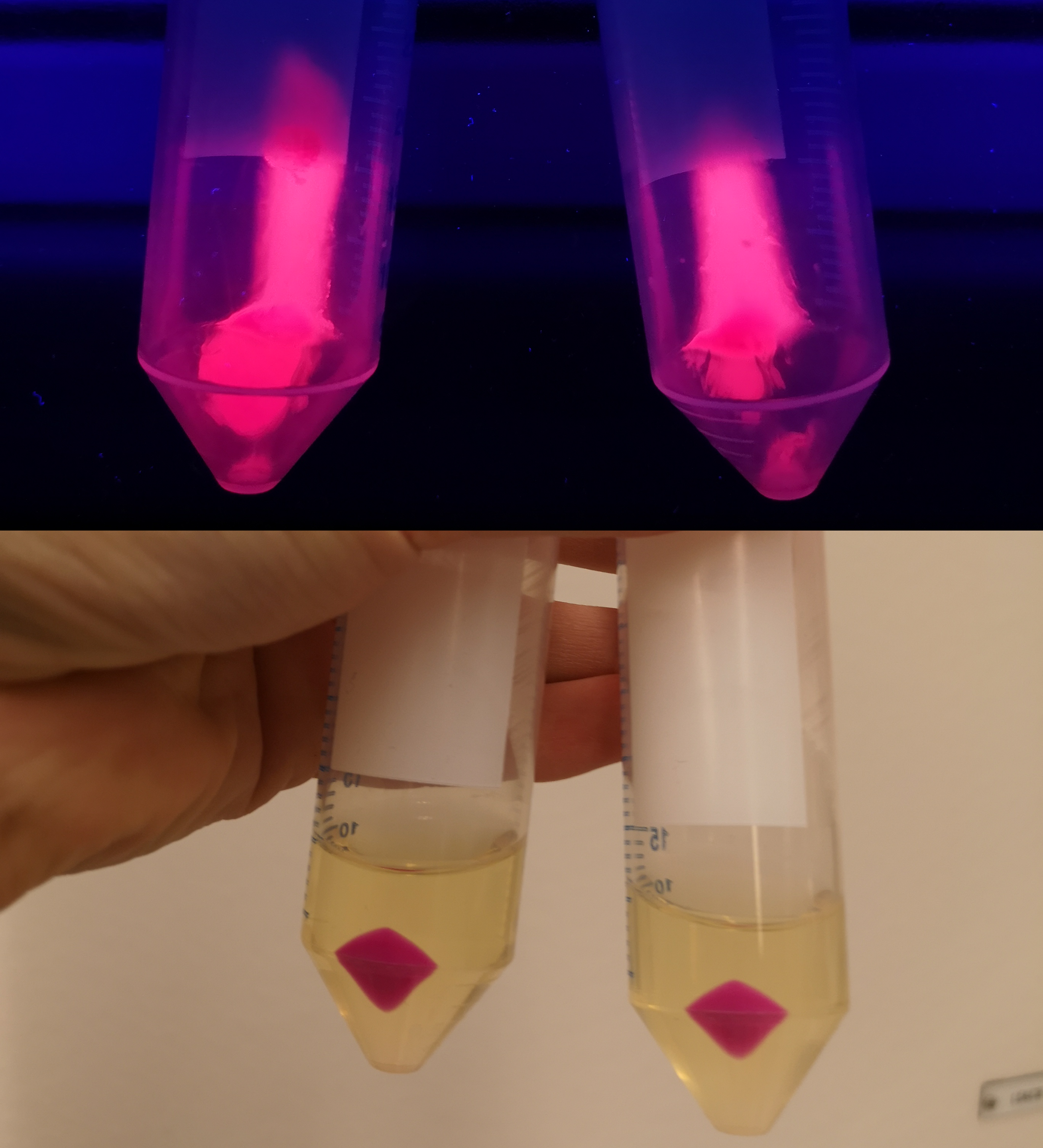
| + | To verify eforReds absorbance and emission, the construct was heat shocked into <i>Escherichia coli</i>, using the strain BL21 (DE3), and grown in a Falcon tube O.N. in 37 °C at 25 µg/mL chloramphenicol. Cotton plugs was used as corks for the Falcon tube. The bacterial solution was compared to a negative control with only BL21 (DE3)(Figure 1, left side) which showed the red color of eforRed in a bacterial solution. Thereafter, the eforRed expressing bacteria was centrifuged at 12 000 g for 10 minutes which displayed a burgundy colour (Figure 1, top-right corner). The pellet was also placed on an UV-table emitting light at 302 nm (Figure 1, down-right corner) and exhibited a pink glowing colour. These experiments verifies eforRed fluorescent effect as well as its absorbance in white light.</p> | ||
| + | |||
<br><br> | <br><br> | ||
| − | + | ||
| − | + | ||
</html>[[Image:T--Linkoping_Sweden--pcons-eforred.jpeg|460px]]<html> | </html>[[Image:T--Linkoping_Sweden--pcons-eforred.jpeg|460px]]<html> | ||
</html>[[Image:T--Linkoping_Sweden--eforred-pellet-photo.png|400px]]<html> | </html>[[Image:T--Linkoping_Sweden--eforred-pellet-photo.png|400px]]<html> | ||
<br><br> | <br><br> | ||
| − | <div class="figurtext"style=font-size: | + | |
| + | <div class="figurtext" style=font-size:90%;><b>Figure 1.</b> The picture to the left depicts a cell culture of BL21 (DE3) pCons-eforRed (left tube) versus a negative control (right tube) with BL21 (DE3). The top right picture displays a pellet of BL21 (DE3) with pCons-eforRed in UV-light. The right picture is a pellet of BL21 (DE3) in white light.</div> | ||
| + | </div> | ||
| + | |||
<br><br> | <br><br> | ||
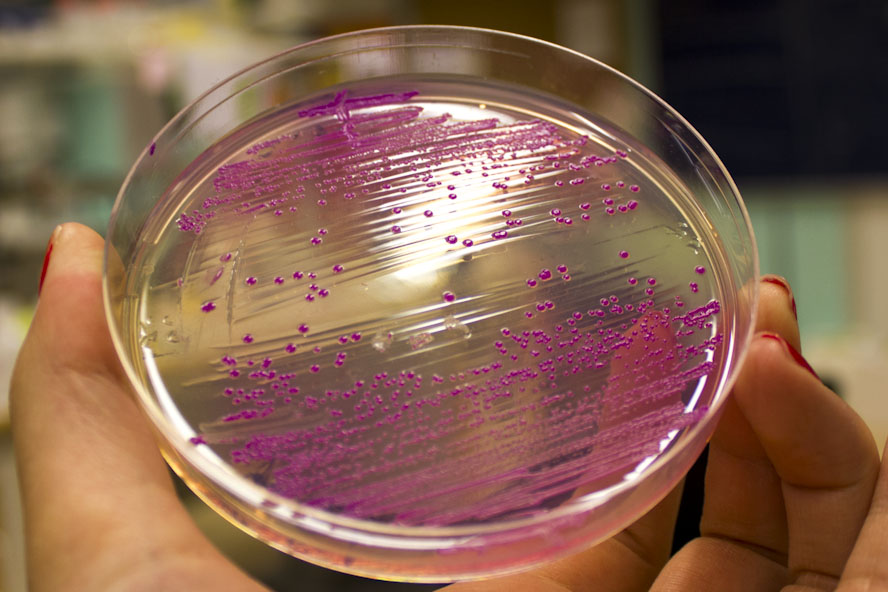
| − | Further characterization was performed in order to demonstrate the absorbance and fluorescence of | + | Further characterization was performed in order to demonstrate the absorbance and fluorescence of eforRed. BL21 (DE3) containing pCons-eforRed were spread on a petri dish containing 25 µg/ml chloramphenicol and was photographed in white light and on an UV-table emitting 302 nm (Figure 2). The results were the same as above, in white light (Figure 2, right) the cultures had a burgundy color and on the UV-table the eforRed expressing bacteria exhibited a pink glowing colour (Figure 2, left). |
| − | + | ||
<br><br> | <br><br> | ||
| + | |||
<div> | <div> | ||
| − | </html>[[Image:T--Linkoping_Sweden--eforred-agar-photo.png|700px | + | </html>[[Image:T--Linkoping_Sweden--eforred-agar-photo.png|700px|left|thumb|<b>Figure 2.</b> Colonies in the same host as previously is presented in white light (right side) and in 302 nm UV-light (left side).]]<html> |
| − | + | ||
| − | + | ||
</div> | </div> | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
<div> | <div> | ||
| − | |||
<p> | <p> | ||
| − | To test the oxygen dependency of the protein production of eforRed in | + | <b style="font-size:120%;">Oxygen dependency</b> |
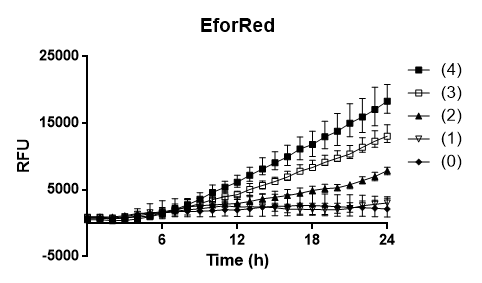
| + | To test the oxygen dependency of the protein production of eforRed in BL21 (DE3), the bacteria containing pCons-eforRed was grown O.N. to 2 OD<sub>600</sub> and diluted to 0.49 OD<sub>600</sub> with LB-miller. The bacteria was placed in a 96-well plate in replicates of 4 with 200 µL in each well. The oxygen access was varied by piercing different numbers of holes (0, 1, 2, 3 and 4) in the plastic film of the 96-well plate. A spectrometry experiment was conducted measuring the fluorescence (excitation 589, emission 609) in 37 °C for 24 hours and the experiment (Figure 3) showed that the access to oxygen effects the folding of eforRed and that 4 holes gave the highest yield.</p> | ||
</div> | </div> | ||
| + | |||
<br> | <br> | ||
| + | |||
<div> | <div> | ||
</html>[[Image:T--Linkoping_Sweden--efffforedd.png|460px]]<html> | </html>[[Image:T--Linkoping_Sweden--efffforedd.png|460px]]<html> | ||
| Line 52: | Line 71: | ||
</div> | </div> | ||
<br><br> | <br><br> | ||
| − | <div | + | <div style=font-size:90%;> |
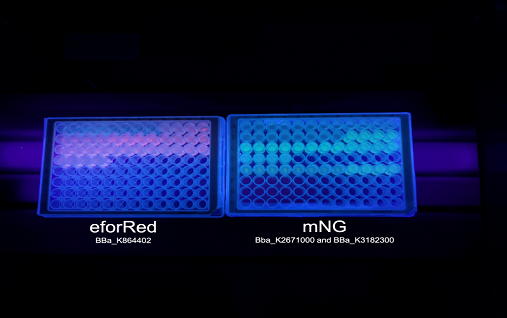
| − | + | <b>Figure 3.</b> To the left is a spectrometry experiment of BL21´s (DE3) protein production of eforRed with varying access to oxygen. The y-axis depicts the relative fluorescence intensity and the x-axis represents the time up to 24 hours. The plate with eforRed can be seen on the right pictures left side and the one on the right side is <i>E.coli</i> BL21 (DE3) with a different expression system used to test a strong green/yellow fluorescent protein. Both plates were illuminated in 302 nm UV-light. | |
| − | + | </div> | |
| + | |||
<br><br> | <br><br> | ||
| + | <p><b style="font-size:120%;">Molecular weight</b><br> | ||
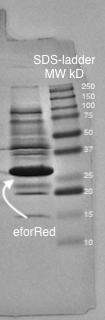
A study of the molecular weight of pCons eforRed expressed in <i>E.coli</i> BL21 (DE3) was done by sonicating the cells and performing a SDS-PAGE electrophoresis on the lysate (<b>Figure 4.</b>). Biorads "Precision Plus Protein Dual Color Standards" was used as the protein ladder in the electrophoresis.<br><br> | A study of the molecular weight of pCons eforRed expressed in <i>E.coli</i> BL21 (DE3) was done by sonicating the cells and performing a SDS-PAGE electrophoresis on the lysate (<b>Figure 4.</b>). Biorads "Precision Plus Protein Dual Color Standards" was used as the protein ladder in the electrophoresis.<br><br> | ||
| + | </p> | ||
</html>[[Image:T--Linkoping_Sweden--eforred_SDS-page.png|100px|left|]]<html> | </html>[[Image:T--Linkoping_Sweden--eforred_SDS-page.png|100px|left|]]<html> | ||
| − | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><div class="figurtext"style=font-size:80%;><b | + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
| + | <div class="figurtext"style=font-size:80%;> | ||
| + | <b>Figure 4.</b> SDS-page of sonicated <I>E.coli</I> BL21 (DE3) lysate with pCons-eforRed. Biorads "Precision Plus Protein Dual Color Standards" was used as the protein ladder. The visible band on the gel lies between 25 and 37 kD which corresponds to the molecular weight of eforRed which is 26.1 kDa. | ||
</div> | </div> | ||
| − | < | + | </html> |
| − | + | <br><br> | |
| − | + | ||
| − | + | ||
| − | + | ||
<!-- --> | <!-- --> | ||
Latest revision as of 16:51, 20 October 2019
J23110-B0034-eforRed
eforRed eforRed is previously described as BBa_K592012. We submitted a functionally active variant with J23110 and B0034.
Usage and Biology
Contribution
Group: Linkoping_Sweden iGEM 2019
Author: Andreas Holmqvist and Leo Juhlin
Summary:
In this contribution, we characterized the visual absorbance and the fluorescence of this construct. We also tested the oxygen dependency of the protein expression in E. coli BL21 (DE3) cells. The molecular weight of eforRed was also examined with an SDS-PAGE.
Documentation:
- the BBa_J23110Constitutive promotor
- the BBa_B0034Ribosome binding site
Fluorescence and absorbance
To verify eforReds absorbance and emission, the construct was heat shocked into Escherichia coli, using the strain BL21 (DE3), and grown in a Falcon tube O.N. in 37 °C at 25 µg/mL chloramphenicol. Cotton plugs was used as corks for the Falcon tube. The bacterial solution was compared to a negative control with only BL21 (DE3)(Figure 1, left side) which showed the red color of eforRed in a bacterial solution. Thereafter, the eforRed expressing bacteria was centrifuged at 12 000 g for 10 minutes which displayed a burgundy colour (Figure 1, top-right corner). The pellet was also placed on an UV-table emitting light at 302 nm (Figure 1, down-right corner) and exhibited a pink glowing colour. These experiments verifies eforRed fluorescent effect as well as its absorbance in white light.

Further characterization was performed in order to demonstrate the absorbance and fluorescence of eforRed. BL21 (DE3) containing pCons-eforRed were spread on a petri dish containing 25 µg/ml chloramphenicol and was photographed in white light and on an UV-table emitting 302 nm (Figure 2). The results were the same as above, in white light (Figure 2, right) the cultures had a burgundy color and on the UV-table the eforRed expressing bacteria exhibited a pink glowing colour (Figure 2, left).
Oxygen dependency To test the oxygen dependency of the protein production of eforRed in BL21 (DE3), the bacteria containing pCons-eforRed was grown O.N. to 2 OD600 and diluted to 0.49 OD600 with LB-miller. The bacteria was placed in a 96-well plate in replicates of 4 with 200 µL in each well. The oxygen access was varied by piercing different numbers of holes (0, 1, 2, 3 and 4) in the plastic film of the 96-well plate. A spectrometry experiment was conducted measuring the fluorescence (excitation 589, emission 609) in 37 °C for 24 hours and the experiment (Figure 3) showed that the access to oxygen effects the folding of eforRed and that 4 holes gave the highest yield.
Molecular weight
A study of the molecular weight of pCons eforRed expressed in E.coli BL21 (DE3) was done by sonicating the cells and performing a SDS-PAGE electrophoresis on the lysate (Figure 4.). Biorads "Precision Plus Protein Dual Color Standards" was used as the protein ladder in the electrophoresis.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Note: This part did not have a reference sequence. A reference sequence has since been added based on the part's documentation; a composite part using the following basic parts: No part name specified with partinfo tag. - BBa_B0034 - BBa_K592012- iGEM HQ