Difference between revisions of "Part:BBa I746909:Experience"
(→Applications of BBa_I746909) |
(→Applications of BBa_I746909) |
||
| (8 intermediate revisions by 3 users not shown) | |||
| Line 4: | Line 4: | ||
===Applications of BBa_I746909=== | ===Applications of BBa_I746909=== | ||
| − | |||
<html> | <html> | ||
| − | <h2> | + | <h2> 2021 Lambert_GA iGEM Team </h2> |
| − | <p> <a href="https:// | + | <p> <a href="https://2021.igem.org/Team:Lambert_GA">2021 Lambert_GA iGEM Team</a> validated this part in our cell-free lysates of BL21 (DE3).<br><br></p> |
</html> | </html> | ||
| + | Lambert_GA iGEM 2021 developed cell-free lysates from E.coli BL21(DE3) competent cells. These lysates were then tested with BBa_I746909 and IPTG induction. Fluorescence was measured three hours after incubating lysate, plasmid BBa_I746909, and master mix at 37ºC. Excitation was measured at 485 nm and emission at 510 nm. The lysate and plasmid BBa_I746909 were tested without IPTG induction and 10mM IPTG. The graph below shows sfGFP expression for our samples. | ||
| + | [[File:CellFreesfGFPExpression.png|thumb|center|500px|<i>Figure 2. The table above shows the cell-free sfGFP expression of blank, lysate+BBa_I746909, and lysate+BBa_I746909+IPTG.</i>]] | ||
| − | |||
| − | |||
| − | + | This validates this part because the T7 RNAP naturally found in the E. coli BL21 strain is able to express the T7 driven sfGFP, and additional IPTG results in additional expression. This also verifies the efficacy of our cell-free lysates. | |
| − | |||
| − | + | <html> | |
| − | + | <h2>2019 iGEM team Gunma Japan</h2> | |
| − | + | <p> <a href="https://2019.igem.org/Team:Gunma">2019 iGEM team Gunma Japan</a>validated this part in BL21(DE3).<br><br></p> | |
| − | + | ||
| − | + | </html> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | We moved our data from here to the main page! | ||
| + | Please check : https://parts.igem.org/Part:BBa_I746909 | ||
| + | Gunma | ||
<html> | <html> | ||
Latest revision as of 17:49, 20 October 2021
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_I746909
2021 Lambert_GA iGEM Team
2021 Lambert_GA iGEM Team validated this part in our cell-free lysates of BL21 (DE3).
This validates this part because the T7 RNAP naturally found in the E. coli BL21 strain is able to express the T7 driven sfGFP, and additional IPTG results in additional expression. This also verifies the efficacy of our cell-free lysates.
2019 iGEM team Gunma Japan
2019 iGEM team Gunma Japanvalidated this part in BL21(DE3).
We moved our data from here to the main page! Please check : https://parts.igem.org/Part:BBa_I746909
Gunma
2018 iGEM team Linkoping Sweden
2018 iGEM team Linkoping Sweden validated this part.
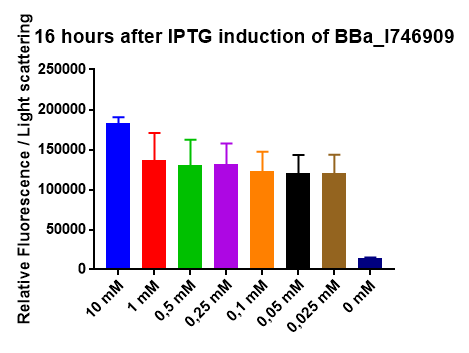
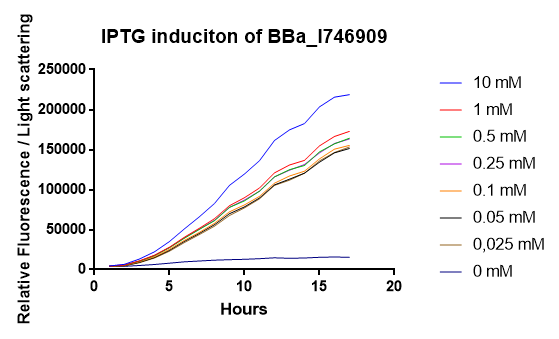
[http://2014.igem.org/Team:Imperial iGEM Team Imperial
[http://2014.igem.org/Team:Aachen iGEM Team Aachen
The iGEM Team Aachen used the biobrick I746909 to test their [http://2014.igem.org/Team:Aachen/Project/2D_Biosensor sensor chip technology].

|
| Testing I746909 in sensor chips I746909 in sensor chips induced with 2µl IPTG and measured with a plate reader. Top chip is not induced, bottom chip is induced with IPTG. |
The biorbick I746909 was also used to test the measurement device [http://2014.igem.org/Team:Aachen/Project/Measurement_Device WatsOn].

|
| Testing I746909 in WatsOn I746909 in sensor chips induced with 2µl IPTG and measured with WatsOn. Left chip is not induced, right chip is induced with IPTG. |
2015 iGEM team Bielefeld-CeBiTec
2015 iGEM team Bielefeld-CeBiTec used this part and improved it by addition of a designed, translation enhancing 5'-untranslated region (5'-UTR; BBa_K1758100). You can find further information at BBa_K1758102).
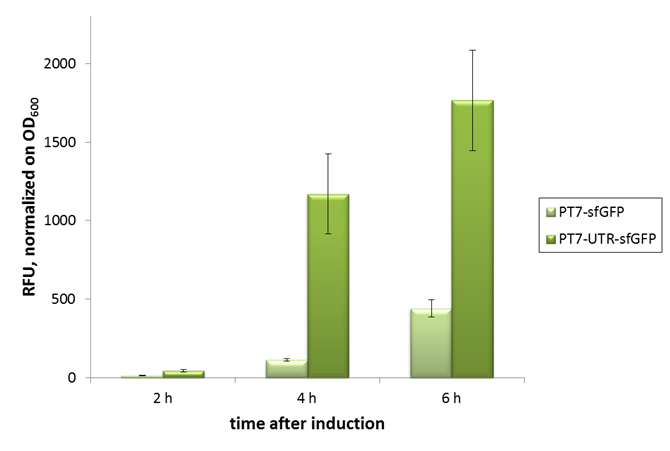
As can be seen in the picture, the difference was observable with the naked eye as well.
[http://2014.igem.org/Team:Imperial iGEM Team Imperial
[http://2014.igem.org/Team:Aachen iGEM Team Aachen
The iGEM Team Aachen used the biobrick I746909 to test their [http://2014.igem.org/Team:Aachen/Project/2D_Biosensor sensor chip technology].

|
| Testing I746909 in sensor chips I746909 in sensor chips induced with 2µl IPTG and measured with a plate reader. Top chip is not induced, bottom chip is induced with IPTG. |
The biorbick I746909 was also used to test the measurement device [http://2014.igem.org/Team:Aachen/Project/Measurement_Device WatsOn].

|
| Testing I746909 in WatsOn I746909 in sensor chips induced with 2µl IPTG and measured with WatsOn. Left chip is not induced, right chip is induced with IPTG. |
User Reviews
UNIQ6360324f68633f57-partinfo-00000008-QINU UNIQ6360324f68633f57-partinfo-00000009-QINU