Difference between revisions of "Part:BBa K115002"
| (7 intermediate revisions by 3 users not shown) | |||
| Line 59: | Line 59: | ||
== Characterized by BNU-China 2019 == | == Characterized by BNU-China 2019 == | ||
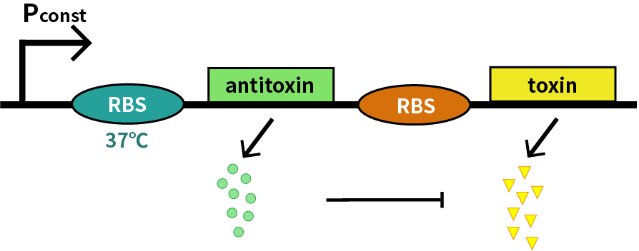
| − | We characterize this RNA thermometer (FourU)(BBa_K115002) | + | We characterize this RNA thermometer (FourU) part (BBa_K115002) by an antitoxin-toxin system, in which the downstream gene encodes for a constantly expressed toxin (relE: BBa_K185000) and the upstream gene encodes for a labile antitoxin (relB: BBa_K185048) under the control of the RNA thermometer (FourU). The expression of antitoxin counteracts lethality of toxin, which otherwise would cause death of bacteria. As a result, we can characterize the behavior of RNA thermometer (FourU) at different temperatures in a cell density-dependent manner in Escherichia coli K-12. |
[[Image:2019 BNU-China BBa K115002 fig1.png| border | center | 400px]]<br> | [[Image:2019 BNU-China BBa K115002 fig1.png| border | center | 400px]]<br> | ||
| Line 77: | Line 77: | ||
2. A strain containing a vector with same backbone is used as control. Experimental groups and control groups are both cultured in 60mL LB-ampicillin (50 ng/µl) medium overnight at 37℃, 200rpm.<br> | 2. A strain containing a vector with same backbone is used as control. Experimental groups and control groups are both cultured in 60mL LB-ampicillin (50 ng/µl) medium overnight at 37℃, 200rpm.<br> | ||
3. Equally divide each group into two flasks, which is 30mL respectively. One of each group is cultured at 27℃, 200rpm and the other at 37℃, 200rpm.<br> | 3. Equally divide each group into two flasks, which is 30mL respectively. One of each group is cultured at 27℃, 200rpm and the other at 37℃, 200rpm.<br> | ||
| − | 4. Extract 5μl samples of each culture system every 6 hours. Diluted all of the samples to | + | 4. Extract 5μl samples of each culture system every 6 hours. Diluted all of the samples to 10^7 times and then spread them on solid LB-ampicillin (50 ng/µl) medium separately.<br> |
| − | 5. Count the number of colonies in 5 | + | 5. Count the number of colonies in 5 cm^2 per plate after cultured for 24 hours at 37℃.<br> |
6. Three repicas are tested in each group.<br> | 6. Three repicas are tested in each group.<br> | ||
| + | |||
| + | <!--Rice iGEM 2019 improvement --> | ||
| + | |||
| + | ==Characterization== | ||
| + | |||
| + | <html> | ||
| + | <p>To characterize this RNA thermometer in vivo, we made an RNA thermometer testing circuit, transformed it into <i>E. coli</i>, and tested the fluorescent output at different temperatures. These graphs show the relative fluorescent intensity of each thermometer between either 25 °C and 30 °C or 25 °C and 37 °C</p> | ||
| + | <img src="https://2019.igem.org/wiki/images/a/a2/T--Rice--RNATcassette.svg" width = 60%> | ||
| + | <p><b>Figure 1.</b>RNA thermometer testing circuit</p> | ||
| + | <img src="https://2019.igem.org/wiki/images/0/05/T--Rice--BBa115002.png" width = 60%> | ||
| + | <p><b>Figure 2.</b> Fluorescence intensity at 25 °C, 30 °C, and 37 °C.</p> | ||
| + | <p>We improved the part BBa_K11502, an RNA thermometer optimized for unfolding at 37 °C. By modifying the nucleotide sequence around the Shine-Dalgarno sequence, we were able to create a new part, <a href="https://parts.igem.org/Part:BBa_K115002">BBa_K3247005</a>, which we called RNA Thermometer NoChill-06, that obtains a much higher change in expression between 25 °C and 37 °C. Please look at the <a href="https://2019.igem.org/Team:Rice/Model" width="100%" margin="10%" height="600px">thermometer design page</a> for more details about our thermometer design process. It's important to note that the Shine Dalgarno sequences used are slightly different. The Tu Delft's team used AGGAGG, while the Rice 2019 iGEM team used AGGAGA.</p> | ||
| + | </html> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 89: | Line 102: | ||
| − | + | ||
| − | + | ||
| + | |||
| + | |||
| + | ==Functional Parameters: Austin_UTexas== | ||
| + | <html> | ||
| + | <body> | ||
<partinfo>BBa_K115002 parameters</partinfo> | <partinfo>BBa_K115002 parameters</partinfo> | ||
| − | < | + | <h3><center>Burden Imposed by this Part:</center></h3> |
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/f/fa/T--Austin_Utexas--no_burden_icon.png" style = "width:160px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: 8.4 ± 17.7% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </body> | ||
| + | </html> | ||
Latest revision as of 22:48, 3 September 2020
RNA thermometer (FourU)
An [http://2008.igem.org/Team:TUDelft/Temperature_analysis#RNA_thermometer RNA thermometer] that can be used for temperature sensitive post-transcriptional regulation which is based on a RNA thermometer from Salmonella Enterica Tyhpy that initiates translation at 37°C.
The image shows the secondary structure of the part after ligation to a protein coding part, as predicted by [http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi RNAfold]. Notice that the 3' end, including the scar and the start codon, does not belong to the part. The blue region is the Shine Dalgarno sequence which is the ribosome binding site. The blue nucleotides are the ones that had to be changed in order to get the desired secondary structure after introduction of the scar.
Free-living microorganisms are frequently exposed to changing environmental conditions. Nutrient availability, osmolarity, pH, temperature and many other parameters are being surveyed constantly. To avoid hazardous consequences, bacteria have established an intricate network of protective mechanisms. Often, regulation of environmentally controlled genes is realized at the transcriptional level by the action of regulatory proteins. However, several post-transcriptional, RNA-based strategies have been discovered recently. It is becoming increasingly clear that certain messenger RNAs (mRNAs) are not simply a substrate for ribosomes but contain control elements that modulate their own expression in a condition-dependent fashion. Structural changes in such sensory RNAs are induced by specific environmental changes. Two principally different classes can be distinguished: cis-acting RNA elements that bear their regulatory potential embedded within the mRNA sequence and trans-acting small, noncoding RNAs that function by base-pairing with complementary mRNA sequences encoded elsewhere in the genome. look more at [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6976.2005.004.x/full here] (edited by TCFSH_Taiwan)
2017 Team SVCE_CHENNAI's characterisation
We characterised the RNA thermometer(FourU) (BBa_K115002) part using GFP to show maximum fluroscence at 37ºC. The part works as expected and maximum level of fluorescence was observed at 37ºC.

The expression of GFP was found to be basal and minimum at 32ºC. A sudden induction in the expression was observed at the temperature 37ºC and it seemed to be gradually decreasing after 42ºC. This behaviour of RNA thermometer(FourU) observed supports the data given by other teams. Thus, the RNA thermometer(FourU) works as expected.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]

2017 Team SVCE_CHENNAI's characterisation
We characterised the RNA thermometer(FourU) (BBa_K115002) part using GFP to show maximum fluroscence at 37ºC. The part works as expected and maximum level of fluorescence was observed at 37ºC.

The expression of GFP was found to be basal and minimum at 32ºC. A sudden induction in the expression was observed at the temperature 37ºC and it seemed to be gradually decreasing after 42ºC. This behaviour of RNA thermometer(FourU) observed supports the data given by other teams. Thus, the RNA thermometer(FourU) works as expected. This is the adaptor

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterized by BNU-China 2019
We characterize this RNA thermometer (FourU) part (BBa_K115002) by an antitoxin-toxin system, in which the downstream gene encodes for a constantly expressed toxin (relE: BBa_K185000) and the upstream gene encodes for a labile antitoxin (relB: BBa_K185048) under the control of the RNA thermometer (FourU). The expression of antitoxin counteracts lethality of toxin, which otherwise would cause death of bacteria. As a result, we can characterize the behavior of RNA thermometer (FourU) at different temperatures in a cell density-dependent manner in Escherichia coli K-12.
In order to measure the function of this RNA thermometer(FourU) at 27℃ and 37℃, we take E. coli introduced with a vector with the same backbone as control group.
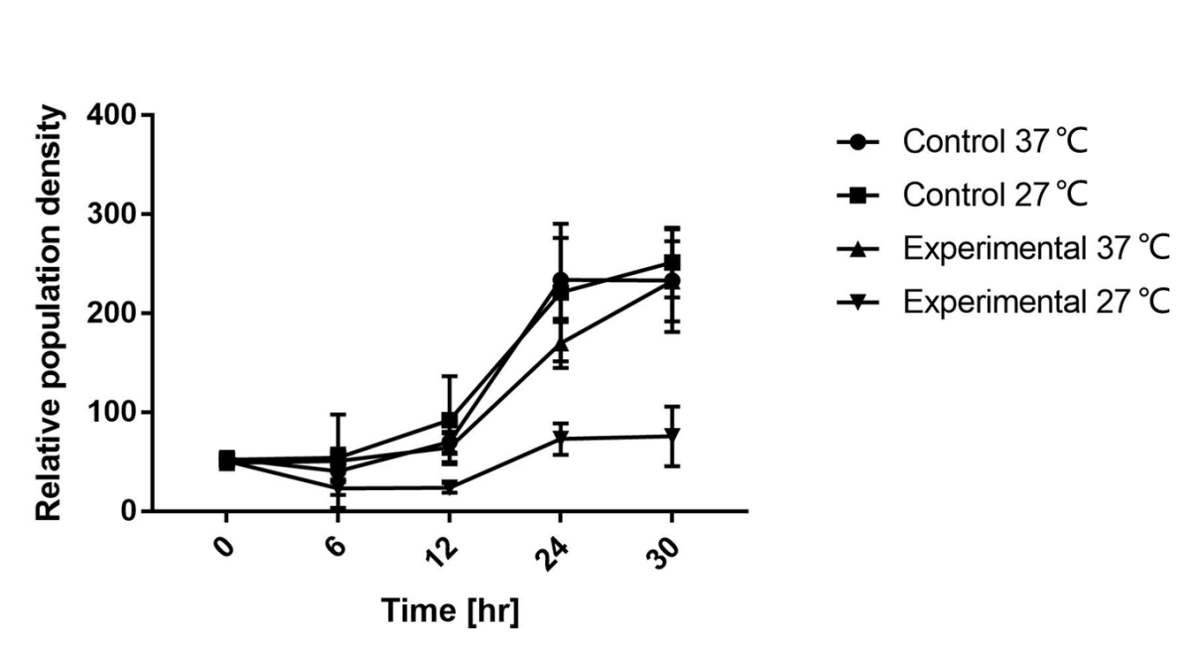
As is shown in Fig.1, there is nearly no difference of the relative population density between control and experimental groups at 37℃, which indicates the RNA thermometer (FourU) conducts expression of downstream gene. However, the population density of experimental group shows a significant decrease compared to control group at 27℃, caused by lack of antitoxin, which indicates the RNA thermometer (FourU) inhibits expression of antitoxin at 27℃.
With properties of the RNA thermometer (FourU), we construct a cold-triggered kill switch to prevent bacteria escaping from human body.
Experimental approach
1. Transform the plasmids into E. coli DH5α competent cells.
2. A strain containing a vector with same backbone is used as control. Experimental groups and control groups are both cultured in 60mL LB-ampicillin (50 ng/µl) medium overnight at 37℃, 200rpm.
3. Equally divide each group into two flasks, which is 30mL respectively. One of each group is cultured at 27℃, 200rpm and the other at 37℃, 200rpm.
4. Extract 5μl samples of each culture system every 6 hours. Diluted all of the samples to 10^7 times and then spread them on solid LB-ampicillin (50 ng/µl) medium separately.
5. Count the number of colonies in 5 cm^2 per plate after cultured for 24 hours at 37℃.
6. Three repicas are tested in each group.
Characterization
To characterize this RNA thermometer in vivo, we made an RNA thermometer testing circuit, transformed it into E. coli, and tested the fluorescent output at different temperatures. These graphs show the relative fluorescent intensity of each thermometer between either 25 °C and 30 °C or 25 °C and 37 °C

Figure 1.RNA thermometer testing circuit

Figure 2. Fluorescence intensity at 25 °C, 30 °C, and 37 °C.
We improved the part BBa_K11502, an RNA thermometer optimized for unfolding at 37 °C. By modifying the nucleotide sequence around the Shine-Dalgarno sequence, we were able to create a new part, BBa_K3247005, which we called RNA Thermometer NoChill-06, that obtains a much higher change in expression between 25 °C and 37 °C. Please look at the thermometer design page for more details about our thermometer design process. It's important to note that the Shine Dalgarno sequences used are slightly different. The Tu Delft's team used AGGAGG, while the Rice 2019 iGEM team used AGGAGA.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.