Difference between revisions of "Part:BBa K1033910"
(→Characterization: NWU-China 2019) |
|||
| (50 intermediate revisions by 4 users not shown) | |||
| Line 17: | Line 17: | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| Line 29: | Line 28: | ||
[[File:T--NWU-China--characterization-BBa_K1033910%28fwyellow%29.png|900px|thumb|none|alt=fwYellow spectum.|Figure 1. fwYellow absorbance spectrum]] | [[File:T--NWU-China--characterization-BBa_K1033910%28fwyellow%29.png|900px|thumb|none|alt=fwYellow spectum.|Figure 1. fwYellow absorbance spectrum]] | ||
| − | ; | + | <br> |
| + | At the same time, fwYellow can emit strong fluorescence under blue light excitation. | ||
| + | [[File:T--NWU-China--Characterization-yellow.jpg|900px|thumb|none|alt=fluorescence of fwYellow.|Figure 1. under blue light excitation, The left one is fwYellow]]<br> | ||
| + | <br> | ||
| + | When we used the microplate reader to measure the excitation light and absorbed light, we found that the spectrum of fwYellow is too narrow. Due to the low precision of the instrument, it is impossible to accurately measure its excitation light and emission light. The excitation light is around 522 nm and the emission is about 536 nm. | ||
| + | |||
| + | ===Characterization: NWU-CHINA-A 2020=== | ||
| + | |||
| + | Last year, our college team NWU-China did not measure the specific values of fwYellow excitation light and emission light accurately. This year, the NWU-CHINA-A team overexpressed fwYellow protein in E.coli and extracted and purified again.<br> | ||
| + | After 16 hours of fermentation, we added IPTG to overexpress this chromoprotein and continued to cultivate for three hours. Then we centrifuged the broth by High speed freezing centrifuge (4℃,8000rpm,15min). Added the lysis solution according to the ratio of the bacteria’s wet weight to the lysis solution 100mg:3ml, and broke the cells in the ultrasonic cell disruptor(500w, work 3s,Stop 5s, 20min),then centrifuged again to get the protein supernatant (4℃,8000rpm,15min). The wavelength of excitation light and emission light was measured by fluorospectro photometer(F-4700).<br> | ||
| + | |||
| + | <html> | ||
| + | <img style="display: block; | ||
| + | width: 90%;height: 140%;" src="https://2020.igem.org/wiki/images/a/a1/T--NWU-CHINA-A--contribution-fw-bottle.jpg"><div>Figure.1 under UV excitation, The right one is fwYellow</div><br> | ||
| + | We scaned the 3D diagrams of samples to read the best conditions: Ex=508nm, Em=510nm.<br> | ||
| + | <img style="display: block; | ||
| + | width: 90%;height: 140%;" src="https://static.igem.org/mediawiki/parts/3/30/T--NWU-CHINA-A--1.png"><div>Figure.2 3D-scan of fwYellow under 400 volt </div><br> | ||
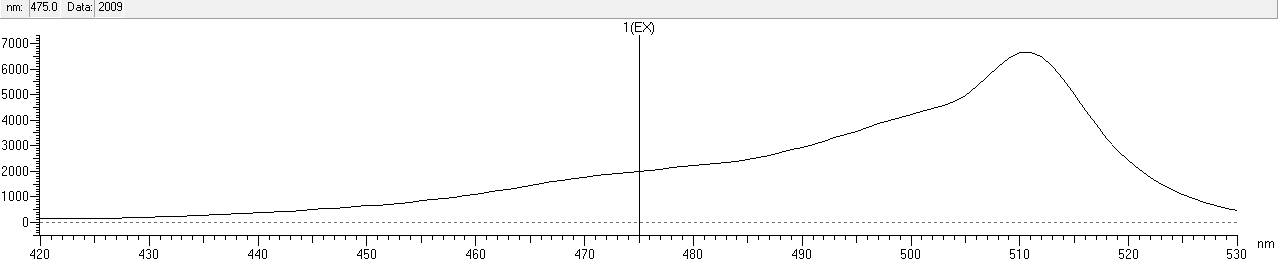
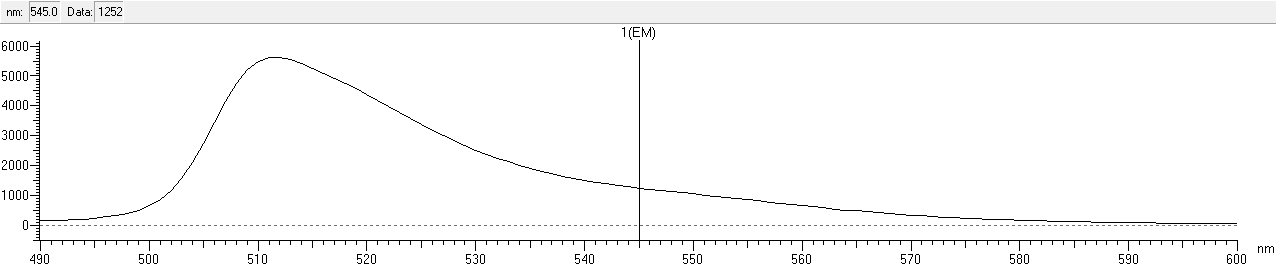
| + | After that, we measured the excitation and emission spectrum of fwYellow. The scanning range of emission spectrum is 420-530nm, and the scanning range of emission spectrum is 490-600nm.<br> | ||
| + | <img style="display: block; | ||
| + | width: 90%;height: 140%;" src="https://2020.igem.org/wiki/images/9/94/T--NWU-CHINA-A--capture_EX-1.png"><div>Figure.3 Excitation spectrum of fwYellow</div><br> | ||
| + | <img style="display: block; | ||
| + | width: 90%;height: 140%;" src="https://2020.igem.org/wiki/images/b/bd/T--NWU-CHINA-A--capture_em-1.png"><div>Figure.4 Emission spectrum of fwYellow </div><br> | ||
| + | </html> | ||
| + | |||
| + | ==Functional Parameters: Austin_UTexas== | ||
| + | <html> | ||
| + | <body> | ||
| + | <partinfo>BBa_K1033910 parameters</partinfo> | ||
| + | <h3><center>Burden Imposed by this Part:</center></h3> | ||
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/f/fa/T--Austin_Utexas--no_burden_icon.png" style = "width:160px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: 0.2 ± 2.4% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | ==Characterization:Worldshaper-NJBIOX 2022== | ||
| + | '''Group: '''iGEM Team Worldshaper-NJBIOX<br> | ||
| + | '''Author:''' Zhiyu Zhang<br> | ||
| + | '''Summary: '''Characterization of the expression of plasmid pET-29a-fwyellow in Escherichia coli strain BL21<br> | ||
| + | |||
| + | Our team obtained the fwyellow gene from iGem team iGEM13_Uppsala and transformed E.coli strain BL21 with plasmid vector pET-29a-fwyellow. We extracted and purified the fwyellow protein expressed by sonication, ultrafiltration, Ni-affinity chromatography, second ultrafiltration, and boiling. The fwyellow produced was analyzed by bio-information analysis (by SWISS-MODEL), electrophoresis, mass spectrum, and chromatography. | ||
| + | |||
| + | ===Experiments Results=== | ||
| + | ====Proof of successful expression==== | ||
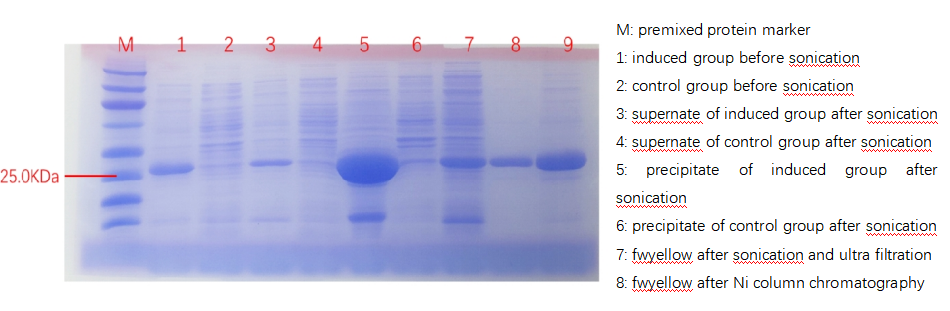
| + | According to bio-information analysis, the estimated molecular mass of fwyellow is 27.9kDa, which matches the electrophoresis results, as shown below. This phenomenon suggests that fwyellow has been successfully synthesized by the transformed bacteria. | ||
| + | [[File:fig1.png|800px|thumb|center|Figure 1. Protein electrophoresis graph]] | ||
| + | |||
| + | '''Yield:''' | ||
| + | We had two groups of transformed E.coli. The final yields of fwyellow were 9.66mg/ml and 6.53mg/ml, respectively. | ||
| + | |||
| + | ====Test of protein characteristics==== | ||
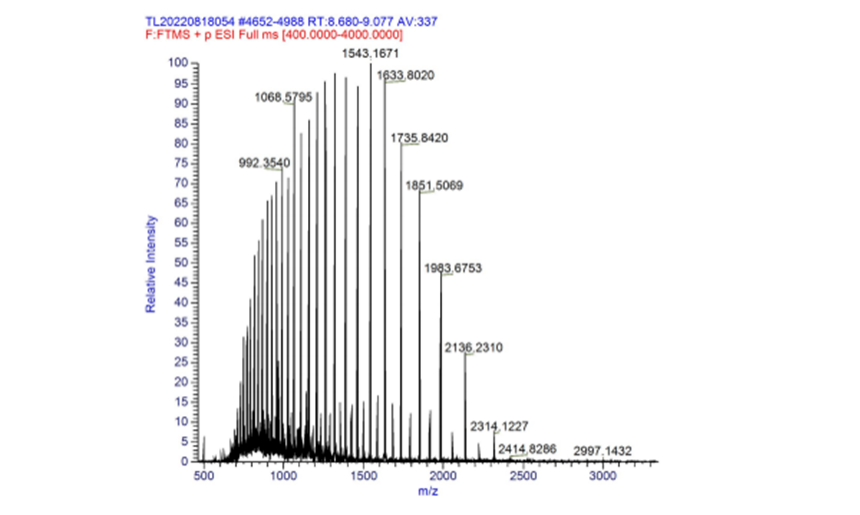
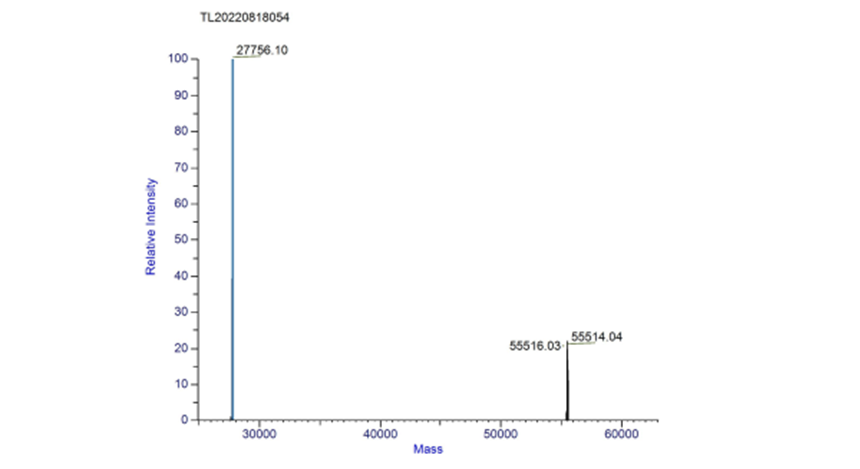
| + | '''Molecular mass:''' | ||
| + | The measured mass of fwyellow is 27756.10 Da and 55514.04 Da (dimer). The primary mass spectrum and deconvolution spectrum graphs are shown below. | ||
| + | [[File:fig2.png|500px|thumb|center|Figure 2. Primary mass spectrum of complete fwyellow molecule]] | ||
| + | [[File:fig3.png|500px|thumb|center|Figure 3. Deconvolution spectrum of complete fwyellow molecule]] | ||
| + | |||
| + | ====Bio-information analysis==== | ||
| + | We submitted the primary amino acid sequence of fwyellow to SWISS-MODEL for bio-information analysis. We acquired additional information, such as the structure model, of the protein. The report is shown below. | ||
| + | [[File:Figure 4. Structure of fwyellow protein.png|500px|thumb|center|Figure 4. Structure of fwyellow protein]] | ||
| + | [[File:fig5.png|800px|thumb|center|Figure 5 Bioinformation analysis report of fwyellow]] | ||
Latest revision as of 06:20, 6 October 2022
fwYellow, yellow chromoprotein
This chromoprotein (also known as FezziwigYFP) naturally exhibits strong color when expressed. Compared to many other chromoproteins, such as amilCP (BBa_K592009), amilGFP (BBa_K592010), spisPink (BBa_K1033932), asPink (BBa_K1033933) and aeBlue (BBa_K864401), the color development is slower. The color is readily observed in both LB or on agar plates after 24-64 hours of incubation.
Usage and Biology
This part is useful as a reporter.
iGEM13_Uppsala: Expression of chromoproteins. The images above show E coli constitutively expressing meffRed BBa_K1033922, aeBlue BBa_K1033929, tsPurple BBa_K1033905, asPink (asCP) BBa_K1033927, fwYellow BBa_K1033910, meffBlue BBa_K1033902, meffRed BBa_K1033922 and scOrange BBa_K1033913.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 109
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 486
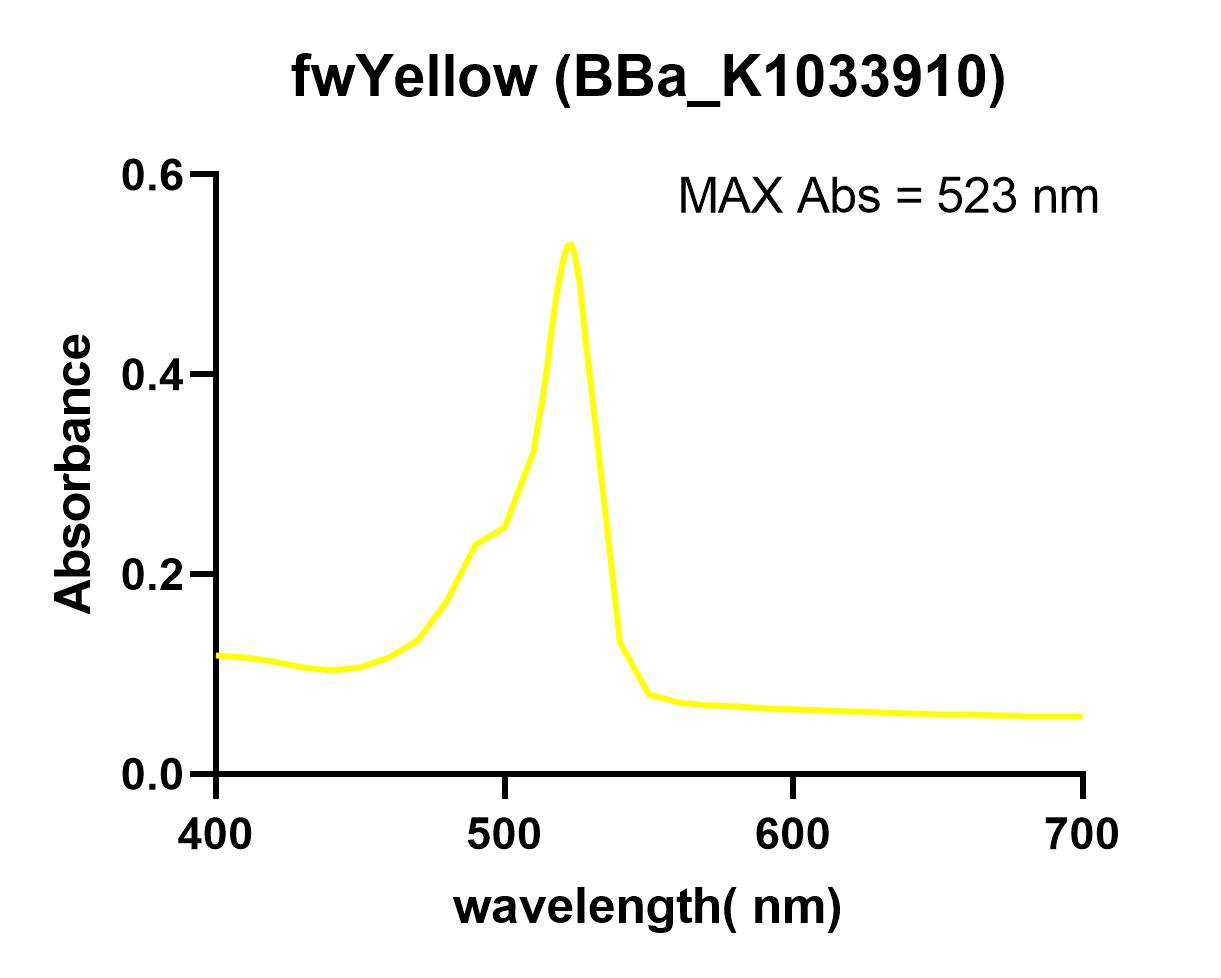
Characterization: NWU-China 2019
The NWU-China team overexpressed fwYellow protein in E. coli and extracted and purified. The maximum absorbance was measured in a 96-well plate using a microplate reader (Synergy 2).
At the same time, fwYellow can emit strong fluorescence under blue light excitation.
When we used the microplate reader to measure the excitation light and absorbed light, we found that the spectrum of fwYellow is too narrow. Due to the low precision of the instrument, it is impossible to accurately measure its excitation light and emission light. The excitation light is around 522 nm and the emission is about 536 nm.
Characterization: NWU-CHINA-A 2020
Last year, our college team NWU-China did not measure the specific values of fwYellow excitation light and emission light accurately. This year, the NWU-CHINA-A team overexpressed fwYellow protein in E.coli and extracted and purified again.
After 16 hours of fermentation, we added IPTG to overexpress this chromoprotein and continued to cultivate for three hours. Then we centrifuged the broth by High speed freezing centrifuge (4℃,8000rpm,15min). Added the lysis solution according to the ratio of the bacteria’s wet weight to the lysis solution 100mg:3ml, and broke the cells in the ultrasonic cell disruptor(500w, work 3s,Stop 5s, 20min),then centrifuged again to get the protein supernatant (4℃,8000rpm,15min). The wavelength of excitation light and emission light was measured by fluorospectro photometer(F-4700).

We scaned the 3D diagrams of samples to read the best conditions: Ex=508nm, Em=510nm.

After that, we measured the excitation and emission spectrum of fwYellow. The scanning range of emission spectrum is 420-530nm, and the scanning range of emission spectrum is 490-600nm.
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
Characterization:Worldshaper-NJBIOX 2022
Group: iGEM Team Worldshaper-NJBIOX
Author: Zhiyu Zhang
Summary: Characterization of the expression of plasmid pET-29a-fwyellow in Escherichia coli strain BL21
Our team obtained the fwyellow gene from iGem team iGEM13_Uppsala and transformed E.coli strain BL21 with plasmid vector pET-29a-fwyellow. We extracted and purified the fwyellow protein expressed by sonication, ultrafiltration, Ni-affinity chromatography, second ultrafiltration, and boiling. The fwyellow produced was analyzed by bio-information analysis (by SWISS-MODEL), electrophoresis, mass spectrum, and chromatography.
Experiments Results
Proof of successful expression
According to bio-information analysis, the estimated molecular mass of fwyellow is 27.9kDa, which matches the electrophoresis results, as shown below. This phenomenon suggests that fwyellow has been successfully synthesized by the transformed bacteria.
Yield: We had two groups of transformed E.coli. The final yields of fwyellow were 9.66mg/ml and 6.53mg/ml, respectively.
Test of protein characteristics
Molecular mass: The measured mass of fwyellow is 27756.10 Da and 55514.04 Da (dimer). The primary mass spectrum and deconvolution spectrum graphs are shown below.
Bio-information analysis
We submitted the primary amino acid sequence of fwyellow to SWISS-MODEL for bio-information analysis. We acquired additional information, such as the structure model, of the protein. The report is shown below.