Difference between revisions of "Part:BBa K3182100"
| (47 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <!-- --> | ||
| + | __TOC__ | ||
| + | |||
| + | <span class='h3bb'><h1>Sequence and Features</h1></span> | ||
| + | <partinfo>BBa_K3182100 SequenceAndFeatures</partinfo> | ||
| + | <br> | ||
| + | <h1>Introduction</h1> | ||
| + | This part is mainly about the pink-white screening method. For more information and characterization of the carbohydrate binding domain (CBDcipA) please see: <partinfo>BBa_K3182108</partinfo>. | ||
| − | |||
<partinfo>BBa_K3182100 short</partinfo> | <partinfo>BBa_K3182100 short</partinfo> | ||
| + | [[File:T--Linkoping_Sweden--fusionproteinillustration.jpg|420px|thumb|right|<b>Figure 1.</b> Mechanism of action for Novosite. The CBD<sub>cipA</sub>-fusion is attached to a polysaccaride material. By adding thrombin from any source the fusion protein will be cleaved and the C-terminal fusion protein will be released into the solution. By changing the fusion protein to an antimicrobial peptide/enzyme, and using the material as a bandage, the peptide/enzyme can be released into a wound by native human thrombin.]] | ||
| − | + | This part consists of a carbohydrate binding domain (CBD) from <i>Clostridium thermocellum (C. thermocellum)</i> cellulose scaffolding protein (CipA). This binding domain is a central part of <i>Clostridium thermocellum's</i> cellusome and has a strong affinity for cellulose. The CBD was fused to another protein using a flexible GS-linker (-GGGGSGGGGS-) in order to attach this complex to a polysaccaride material. A thrombin cleavage site (-LVPRGS-) was added to the end of the linker and its breakage will leave a glycine and serine attached to the N-terminal of the fusion protein. The main mechanism of iGEM19 Linköping's project can be seen in Figure 1. | |
| − | + | <h3>Protease site and use</h3> | |
| + | The thrombin site was added to enable the ability to release the fusion protein down into skin wounds. Thanks to our integrated human practice we learned that infections span much deeper into wounds that we thought. Simply attaching the CBD-fusion protein to a carbohydrate material would not enable the fusion protein to reach far into the wound. The thrombin site was also chosen because of thrombin's endogenous existence in humans. | ||
| + | |||
| + | <h3>Assembly compabilities</h3> | ||
| + | An internal BamHI recognition sequence (RS) has been added to enable interchangeable fusion proteins to the CBD. BamHI was chosen because its RS codes for glycine and serine, fitting it to the end of the thrombin site. It is also a cost-effective enzyme and is unaffected by methylated DNA. BamHI is a part of the RFC21 standard. | ||
| − | |||
<br> | <br> | ||
| − | |||
| − | + | <h2>CBD<sub>cipA</sub> crystal structure</h2> | |
| + | [[File:T--Linkoping_Sweden--rotatingcbdanimationloop.gif|420px|thumb|left|<b>Figure 2.</b> Crystal structure of CBD<sub>cipA</sub> with a resolution of 1.75 Å which were solved by [http://www.ncbi.nlm.nih.gov/pmc/PMC452321 Tormo et al. 1989]. PDB code 1NBC. In red from the left, W118, R112, D56, H57 and Y67, thought to be the surface which interacts strongly with polysaccarides.]] | ||
| − | < | + | <h3>Important molecular faces</h3> |
| + | CBD<sub>cipA</sub> is composed of a nine-stranded beta sandwich with a jelly roll topology and binds a calcium ion, which can be seen in Figure 2. It further contains conserved residues exposed on the surface which map into two clear surfaces on each side of the molecule. One of the faces mainly contains planar strips of aromatic and polar residues which may be the carbohydrate binding part. Further aspects are unknown and unique to this CBD such as the other conserved residues which are contained in a groove. | ||
| − | + | <h3>Carbohydrate binding domain specificity</h3> | |
| + | Since the CBD is from the cellusome of C. thermocellum some research labeled it a cellulose binding domain. However, iGEM19 Linköping noticed that this domain could also bind to different sources of polysaccaride materials. This serves as a domain for iGEM19 Linköpings modular bandage, where the polysaccaride material can be exchanged for other/similar materials and not exclusively cellulose. | ||
| − | [[ | + | <h3>The choice of carbohydrate binding domain</h3> |
| + | iGEM Linköping 2019 chose CBD<sub>cipA</sub> due to the fact that many other iGEM teams had explored the possibilities of this domain. Our basic design was influenced by [http://2014.igem.org/Team:Imperial iGEM14 Imperial], [http://2015.igem.org/Team:edinburgh iGEM15 Edinburgh] and [http://2018.igem.org/Team:ecuador iGEM18 Ecuador]. Purification and where to place the fusion protein (N- or C-terminal) was determined by studying the former projects. CBD<sub>cipA</sub> also originates from a thermophilic bacteria which further increases the domain's applications. | ||
| + | <br><br> | ||
| + | <h2>Expression system</h2> | ||
| + | The part has a strong expression with a T7-RNA-polymerase promotor (<partinfo>BBa_I719005</partinfo>), seen in Figure 3, as well as a 5'-UTR (<partinfo>BBa_K1758100</partinfo>) region which has been shown to further increase expression in <i>Escherichia coli</i> (<i>E. coli</i>) (<partinfo>BBa_K1758106</partinfo>), ([http://www.ncbi.nlm.nih.gov/pubmed/2676996 Olins et al. 1989]), ([http://www.ncbi.nlm.nih.gov/pubmed/23927491 Takahashi et al. 2013]). | ||
| − | + | [[File:T--Linkoping_Sweden--expression.png|900px|thumb|center|<b>Figure 3.</b> Benchling screenshot of the expression system. The T7-RNA-polymerase promotor is followed by a T7 g10 leader sequence which enhances the binding to the 16S ribosomal RNA. After the leader sequence a poly A spacer is found, which has been shown to increase translation in vitro. Before the start codon a strong RBS, g10-L, followed by an AT-rich spacer can be seen, which will slightly increase translation of the following gene.]] | |
| − | + | ||
| − | + | <h3>Theoretical usage of this part</h3> | |
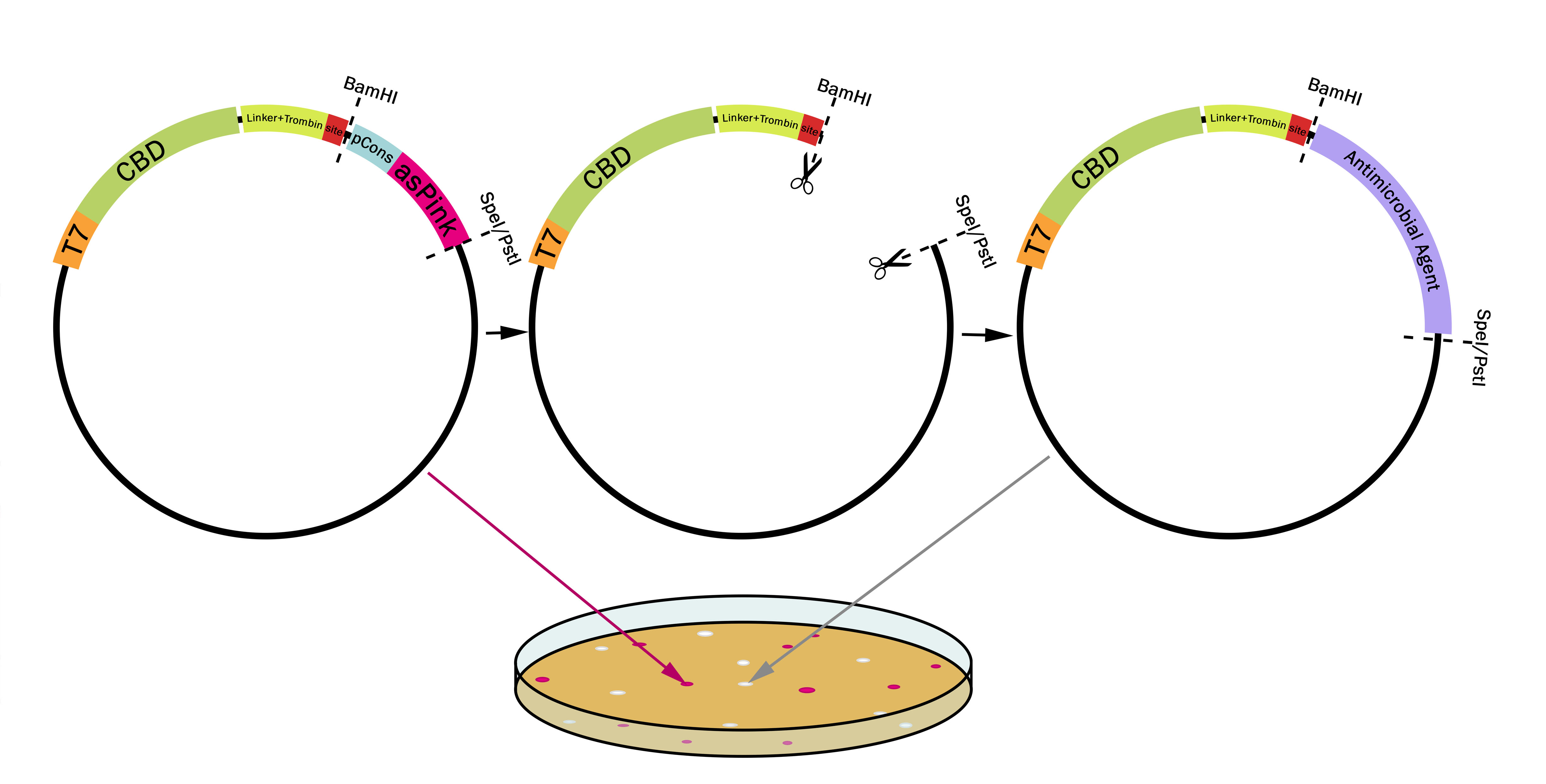
| + | This part utilizes a pCons-AsPink dropout enabling colour-screening for positive colonies. Using BamHI and PstI or SpeI on both these parts assembled in pSB1C3 (or vector of choice) and the insert of choice will yield a fusion protein between CBDcipA and the insert. The fusion protein can later be cleaved with thrombin to generate two separate proteins; the CBD domain and the C-terminally fused protein. The C-terminal protein will have one glycine and one serine added to the N-terminal of the protein. | ||
| − | [[File:T--Linkoping_Sweden-- | + | [[File:T--Linkoping_Sweden--plasmidpstispei.png|800px|thumb|center|<b>Figure 4.</b> The first plasmid (left) contains pCons-AsPink which results in pink colonies. When BamHI is used together with either SpeI or PstI pCons-AsPink is cut out (middle plasmid) and replaced with a compatible biobrick such as an antimicrobial agent (right plasmid). The colonies will then be white due to pCons-AsPink being cleaved from the plasmid. This results in a "pink-white screening". ]] |
| − | + | <h2>Examples of biobricks assembled with this method</h2> | |
| + | <b>This method was used to assemble</b>: <partinfo>BBa_K3182103</partinfo>, <partinfo>BBa_K3182104</partinfo>, <partinfo>BBa_K3182105</partinfo>, <partinfo>BBa_K3182106</partinfo>, <partinfo>BBa_K3182107</partinfo>, <partinfo>BBa_K3182108</partinfo> respectively into pSB1C3. However, as we quickly noticed, pSB1C3 was not able to replicate in <i>Vibrio natriegens</i> (Vmax). After countless failed transformation attempts, all the above parts were also assembled into a modified pUC19 vector using the pink-white screening method. The pUC19 variants were transformed without any problems into <i>V. natriegens</i> (Vmax). A total of 13 fusion proteins were successfully ligated using this method. | ||
| − | |||
| − | + | <h2>Usage and Biology</h2> | |
| + | <h3>Using Pink-White screening</h3> | ||
| + | When using the pink-white screening method, with the goal to fuse any gene to the CBD<sub>cipA</sub>, utilize the BamHI recognition sequence in the 5'-end. This will fuse the gene in frame with CBD<sub>cipA</sub>. The biobrick suffix can be used in the 3'-end. Cut the vector and insert with BamHI and PstI (SpeI also works), remove enzymes and mix. There is no need for gel purification. The use of very high molar ratios might not yield any pink colonies at all. A molar ratio insert to vector of 7-20:1 will yield some pink colonies. Transform the host (BL21 (DE3) for quickest results) and incubate at 37 °C overnight. If the color is weak or can not be seen, incubate in 24-37 °C for an additional 16-24 hours. | ||
| − | + | [[File:T--Linkoping_Sweden--pinkwhite121324.jpeg|700px|thumb|center|<b>Figure 5.</b> (A,B) <i>E. coli</i> (BL21) cells used for pink-white screening after 16 hours in 37 °C. <partinfo>BBa_K3182100</partinfo> was cut with BamHI and PstI to remove pCons-AsPink and <partinfo>BBa_K3182006</partinfo> (Magainin 2) or <partinfo>BBa_K3182104</partinfo> (CHAP) was ligated into the plasmid. The white colonies indicate a successful ligation and the pink indicate an unsuccessful ligation. (C) An agarose gel (1.3 %, TAE) with the biobrick <partinfo>BBa_K3182108</partinfo> (CBD-sfGFP). Lane 1-3 contains three white colonies of CBD-sfGFP DNA amplified from pSB1C3 with VR and VF2. All fragment showed their expected size (1.2 kb). Lane 4 contains a 1 kb Plus DNA Ladder by New England Biolabs.]] | |
| + | |||
| + | <h3>Results using Pink-white screening</h3> | ||
| + | Colonies from the pink-white screening were later sent for sequencing. The results concluded that all the white colonies contained correctly ligated plasmids. The only colony-screen assembly attempt can be seen in Figure 5C, where all colonies screened showed the correct fragments. Therefore, in this attempt, all non-pink colonies showed green fluorescence (<partinfo>BBa_K3182108</partinfo>, sfGFP). After the results from Figure 5C had been sequenced, no more colony screenings had to be done. All white colonies sent to sequencing contained the correctly assembled fusion proteins. | ||
<!-- --> | <!-- --> | ||
| − | |||
| − | |||
Latest revision as of 14:37, 20 October 2019
Contents
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 592
Illegal NheI site found at 615 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 580
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Introduction
This part is mainly about the pink-white screening method. For more information and characterization of the carbohydrate binding domain (CBDcipA) please see: BBa_K3182108.
pT7-CBDcipA-pCons-AsPink

This part consists of a carbohydrate binding domain (CBD) from Clostridium thermocellum (C. thermocellum) cellulose scaffolding protein (CipA). This binding domain is a central part of Clostridium thermocellum's cellusome and has a strong affinity for cellulose. The CBD was fused to another protein using a flexible GS-linker (-GGGGSGGGGS-) in order to attach this complex to a polysaccaride material. A thrombin cleavage site (-LVPRGS-) was added to the end of the linker and its breakage will leave a glycine and serine attached to the N-terminal of the fusion protein. The main mechanism of iGEM19 Linköping's project can be seen in Figure 1.
Protease site and use
The thrombin site was added to enable the ability to release the fusion protein down into skin wounds. Thanks to our integrated human practice we learned that infections span much deeper into wounds that we thought. Simply attaching the CBD-fusion protein to a carbohydrate material would not enable the fusion protein to reach far into the wound. The thrombin site was also chosen because of thrombin's endogenous existence in humans.
Assembly compabilities
An internal BamHI recognition sequence (RS) has been added to enable interchangeable fusion proteins to the CBD. BamHI was chosen because its RS codes for glycine and serine, fitting it to the end of the thrombin site. It is also a cost-effective enzyme and is unaffected by methylated DNA. BamHI is a part of the RFC21 standard.
CBDcipA crystal structure
Important molecular faces
CBDcipA is composed of a nine-stranded beta sandwich with a jelly roll topology and binds a calcium ion, which can be seen in Figure 2. It further contains conserved residues exposed on the surface which map into two clear surfaces on each side of the molecule. One of the faces mainly contains planar strips of aromatic and polar residues which may be the carbohydrate binding part. Further aspects are unknown and unique to this CBD such as the other conserved residues which are contained in a groove.
Carbohydrate binding domain specificity
Since the CBD is from the cellusome of C. thermocellum some research labeled it a cellulose binding domain. However, iGEM19 Linköping noticed that this domain could also bind to different sources of polysaccaride materials. This serves as a domain for iGEM19 Linköpings modular bandage, where the polysaccaride material can be exchanged for other/similar materials and not exclusively cellulose.
The choice of carbohydrate binding domain
iGEM Linköping 2019 chose CBDcipA due to the fact that many other iGEM teams had explored the possibilities of this domain. Our basic design was influenced by [http://2014.igem.org/Team:Imperial iGEM14 Imperial], [http://2015.igem.org/Team:edinburgh iGEM15 Edinburgh] and [http://2018.igem.org/Team:ecuador iGEM18 Ecuador]. Purification and where to place the fusion protein (N- or C-terminal) was determined by studying the former projects. CBDcipA also originates from a thermophilic bacteria which further increases the domain's applications.
Expression system
The part has a strong expression with a T7-RNA-polymerase promotor (BBa_I719005), seen in Figure 3, as well as a 5'-UTR (BBa_K1758100) region which has been shown to further increase expression in Escherichia coli (E. coli) (BBa_K1758106), ([http://www.ncbi.nlm.nih.gov/pubmed/2676996 Olins et al. 1989]), ([http://www.ncbi.nlm.nih.gov/pubmed/23927491 Takahashi et al. 2013]).

Theoretical usage of this part
This part utilizes a pCons-AsPink dropout enabling colour-screening for positive colonies. Using BamHI and PstI or SpeI on both these parts assembled in pSB1C3 (or vector of choice) and the insert of choice will yield a fusion protein between CBDcipA and the insert. The fusion protein can later be cleaved with thrombin to generate two separate proteins; the CBD domain and the C-terminally fused protein. The C-terminal protein will have one glycine and one serine added to the N-terminal of the protein.
Examples of biobricks assembled with this method
This method was used to assemble: BBa_K3182103, BBa_K3182104, BBa_K3182105, BBa_K3182106, BBa_K3182107, BBa_K3182108 respectively into pSB1C3. However, as we quickly noticed, pSB1C3 was not able to replicate in Vibrio natriegens (Vmax). After countless failed transformation attempts, all the above parts were also assembled into a modified pUC19 vector using the pink-white screening method. The pUC19 variants were transformed without any problems into V. natriegens (Vmax). A total of 13 fusion proteins were successfully ligated using this method.
Usage and Biology
Using Pink-White screening
When using the pink-white screening method, with the goal to fuse any gene to the CBDcipA, utilize the BamHI recognition sequence in the 5'-end. This will fuse the gene in frame with CBDcipA. The biobrick suffix can be used in the 3'-end. Cut the vector and insert with BamHI and PstI (SpeI also works), remove enzymes and mix. There is no need for gel purification. The use of very high molar ratios might not yield any pink colonies at all. A molar ratio insert to vector of 7-20:1 will yield some pink colonies. Transform the host (BL21 (DE3) for quickest results) and incubate at 37 °C overnight. If the color is weak or can not be seen, incubate in 24-37 °C for an additional 16-24 hours.

Results using Pink-white screening
Colonies from the pink-white screening were later sent for sequencing. The results concluded that all the white colonies contained correctly ligated plasmids. The only colony-screen assembly attempt can be seen in Figure 5C, where all colonies screened showed the correct fragments. Therefore, in this attempt, all non-pink colonies showed green fluorescence (BBa_K3182108, sfGFP). After the results from Figure 5C had been sequenced, no more colony screenings had to be done. All white colonies sent to sequencing contained the correctly assembled fusion proteins.

