Difference between revisions of "Part:BBa K2549038"
m |
(→Our characterization) |
||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 12: | Line 12: | ||
<!-- Add more about the biology of this part here --> | <!-- Add more about the biology of this part here --> | ||
===Biology=== | ===Biology=== | ||
| − | ===== | + | |
| − | + | ||
| + | =====Our characterization===== | ||
| + | [[File:AND-test.png|none|480px|thumb|'''CfaN intein-based AND gate.''' A degradable EGFP (d2EGFP) is produced downstream the promoter of the Combiner to indicate the output (CusP) strength. DBD, DNA binding domain which is zinc finger in our assay. AD, activating-form transcriptional domain, was VP64. RE, responsive elements. RFI, relative fluorescence intensity (comparing before and after activation). More details please visit http://2018.igem.org/Team:Fudan/Results and http://2018.igem.org/Team:Fudan/Measurement .]] | ||
| + | |||
| + | We show that when VP64-ZF21.16N-CfaN co-expressed with CfaC-ZF21.16C-NLS, the expression level of d2EGFP is relatively turned up due to the formation of VP64-ZF21.16 after auto-plicing and ligation. The signal-noise-ratio is still not optimal, and we are improving the design by switching the split position as well as adding linkers. | ||
| + | |||
| + | [[File:NAND-test.png|none|480px|thumb|'''CfaN intein-based NAND gate.''' A degradable EGFP (d2EGFP) is produced downstream the promoter of the Combiner to indicate the output (CusP) strength. DBD, DNA binding domain which is zinc finger in our assay. SD, silencing-form transcriptional domain, was KRAB. RE, responsive elements. RFI, relative fluorescence intensity (comparing before and after activation).]] | ||
| + | |||
| + | We show that when KRAB-ZF21.16N-CfaN co-expressed with CfaC-ZF21.16C-NLS, the expression level of d2EGFP is relatively turned down due to the formation of KRAB-ZF21.16 after auto-plicing and ligation. | ||
| + | |||
| + | The signal-noise-ratio is ok when both present. However, we are not sure why CfaC-ZF21.16C-NLS alone has effect. We have formed a couple of hypothesis and in the process of optimizing this gate. Please note [[Part:BBa_K2549036]] contains a transcriptional activation domain VP64, while [[Part:BBa_K2549037]] has a transcriptional repression domain KRAB, both need to ligate with CfaC-ZF21.16C-NLS ([[Part:BBa_K2549038]]) before functional. | ||
=====Boolean logic gates via split zinc finger-based transcription factors===== | =====Boolean logic gates via split zinc finger-based transcription factors===== | ||
| − | Lohmueller JJ et al have demonstrated the split ZF-TF reconstitution process. | + | Lohmueller JJ et al have demonstrated the split ZF-TF reconstitution process. Please note that we used Cfa split intein ([[Part:BBa_K2549009]] and [[Part:BBa_K2549010]]) but not dnaB reported below. |
[[File:ZF-TF.jpg|none|400px|thumb|Lohmueller JJ et al demonstrated: ''After expression, the two split ZF-intein fragments bind together and undergo protein splicing to cleave away intein fragments and reconstitute the full ZF activator leading to activation of the BCR_ABL reporter.'']] | [[File:ZF-TF.jpg|none|400px|thumb|Lohmueller JJ et al demonstrated: ''After expression, the two split ZF-intein fragments bind together and undergo protein splicing to cleave away intein fragments and reconstitute the full ZF activator leading to activation of the BCR_ABL reporter.'']] | ||
| Line 23: | Line 33: | ||
[[File:zfNAND.jpeg|none|400px|thumb|Lohmueller JJ et al demonstrated: ''For NAND gates, the computational module splices a ZF repressor, and the logical operation is computed as TRUE as long as both inputs are not present together. For the response data shown BCR_ABL-1:GCN4 repressor split fragments were used and the response promoter contains 6 copies of the BCR_ABL target site. CFP expression was measured by flow cytometry and expressed as fold change over an off-target expression control.'']] | [[File:zfNAND.jpeg|none|400px|thumb|Lohmueller JJ et al demonstrated: ''For NAND gates, the computational module splices a ZF repressor, and the logical operation is computed as TRUE as long as both inputs are not present together. For the response data shown BCR_ABL-1:GCN4 repressor split fragments were used and the response promoter contains 6 copies of the BCR_ABL target site. CFP expression was measured by flow cytometry and expressed as fold change over an off-target expression control.'']] | ||
| − | |||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
| Line 29: | Line 38: | ||
<partinfo>BBa_K2549038 parameters</partinfo> | <partinfo>BBa_K2549038 parameters</partinfo> | ||
--> | --> | ||
| + | |||
===References=== | ===References=== | ||
Latest revision as of 21:36, 17 October 2018
CfaC-ZF21.16C-NLS
This part is one of the downstream elements of our amplifier. It was constructed by fusing CfaC (Part:BBa_K2549010), ZF21.16C (Part:BBa_K2549012) and NLS (Part:BBa_K2549054), from N terminal to C terminal. CfaC is the C-terminal fragment of Cfa which is a consensus sequence from an alignment of 73 naturally occurring DnaE inteins that are predicted to have fast splicing rates. ZF21.16C is the C-terminal fragment of the zinc finger whose recognition helices for three-finger arrays are substituted by the reported synthetic zinc finger 21.16 residues on the basis of the BCR_ABL-1 artificial zinc finger[1]. NLS is a short nuclear location sequence from SV40 large T antigen. When coexpressed with VP64-ZF21.16N-CfaN (Part:BBa_K2549036) in the same cell, both fusions will be produced and a transcription activating function will be executed. Also, when coexpressed with KRAB-ZF21.16N-CfaN (Part:BBa_K2549037) in the same cell, both fusions will be produced formed and a transcription repressing function will be executed.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 91
Illegal SapI.rc site found at 21
Biology
Our characterization
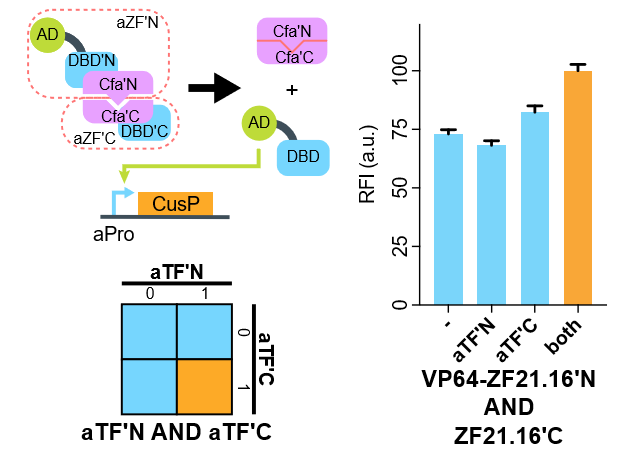
We show that when VP64-ZF21.16N-CfaN co-expressed with CfaC-ZF21.16C-NLS, the expression level of d2EGFP is relatively turned up due to the formation of VP64-ZF21.16 after auto-plicing and ligation. The signal-noise-ratio is still not optimal, and we are improving the design by switching the split position as well as adding linkers.
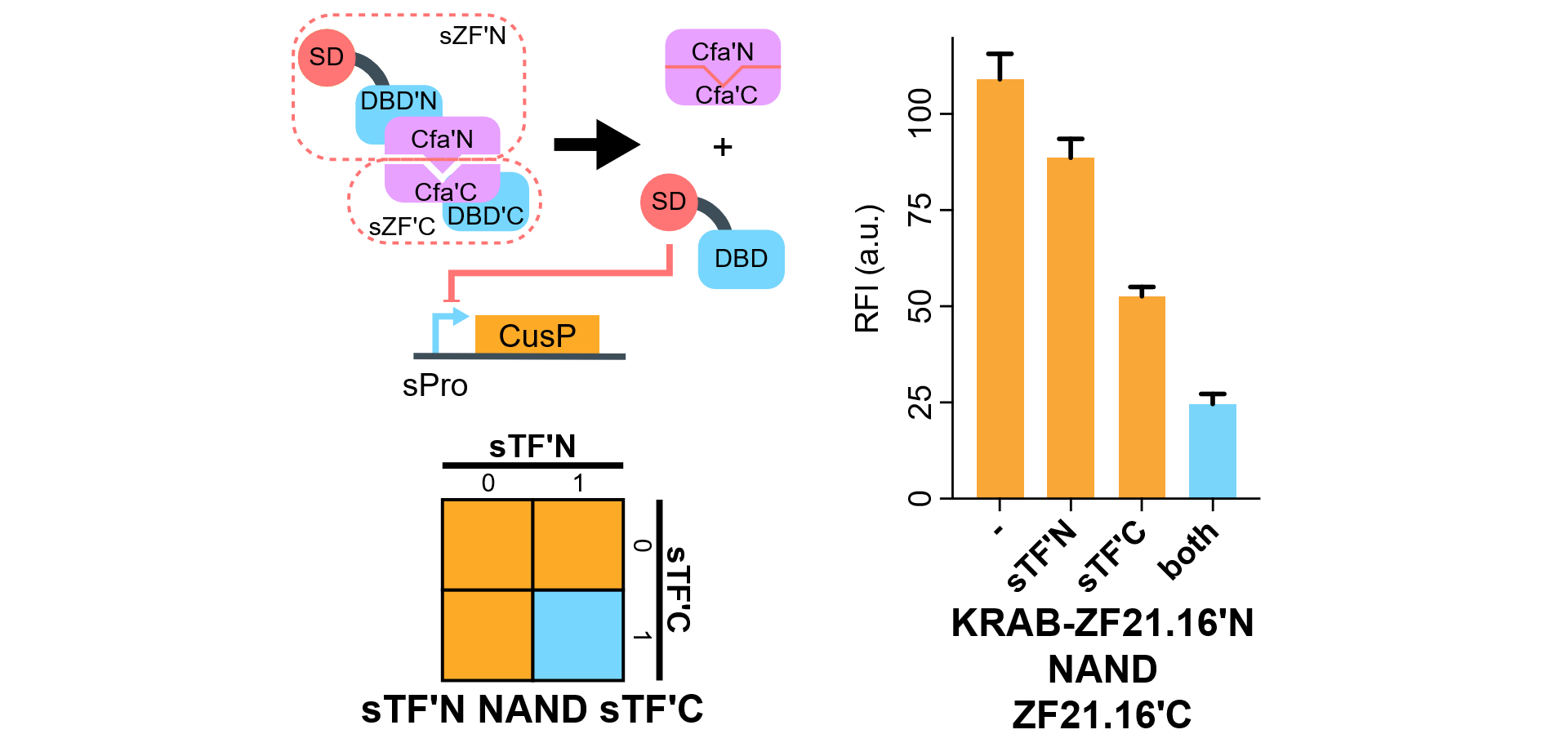
We show that when KRAB-ZF21.16N-CfaN co-expressed with CfaC-ZF21.16C-NLS, the expression level of d2EGFP is relatively turned down due to the formation of KRAB-ZF21.16 after auto-plicing and ligation.
The signal-noise-ratio is ok when both present. However, we are not sure why CfaC-ZF21.16C-NLS alone has effect. We have formed a couple of hypothesis and in the process of optimizing this gate. Please note Part:BBa_K2549036 contains a transcriptional activation domain VP64, while Part:BBa_K2549037 has a transcriptional repression domain KRAB, both need to ligate with CfaC-ZF21.16C-NLS (Part:BBa_K2549038) before functional.
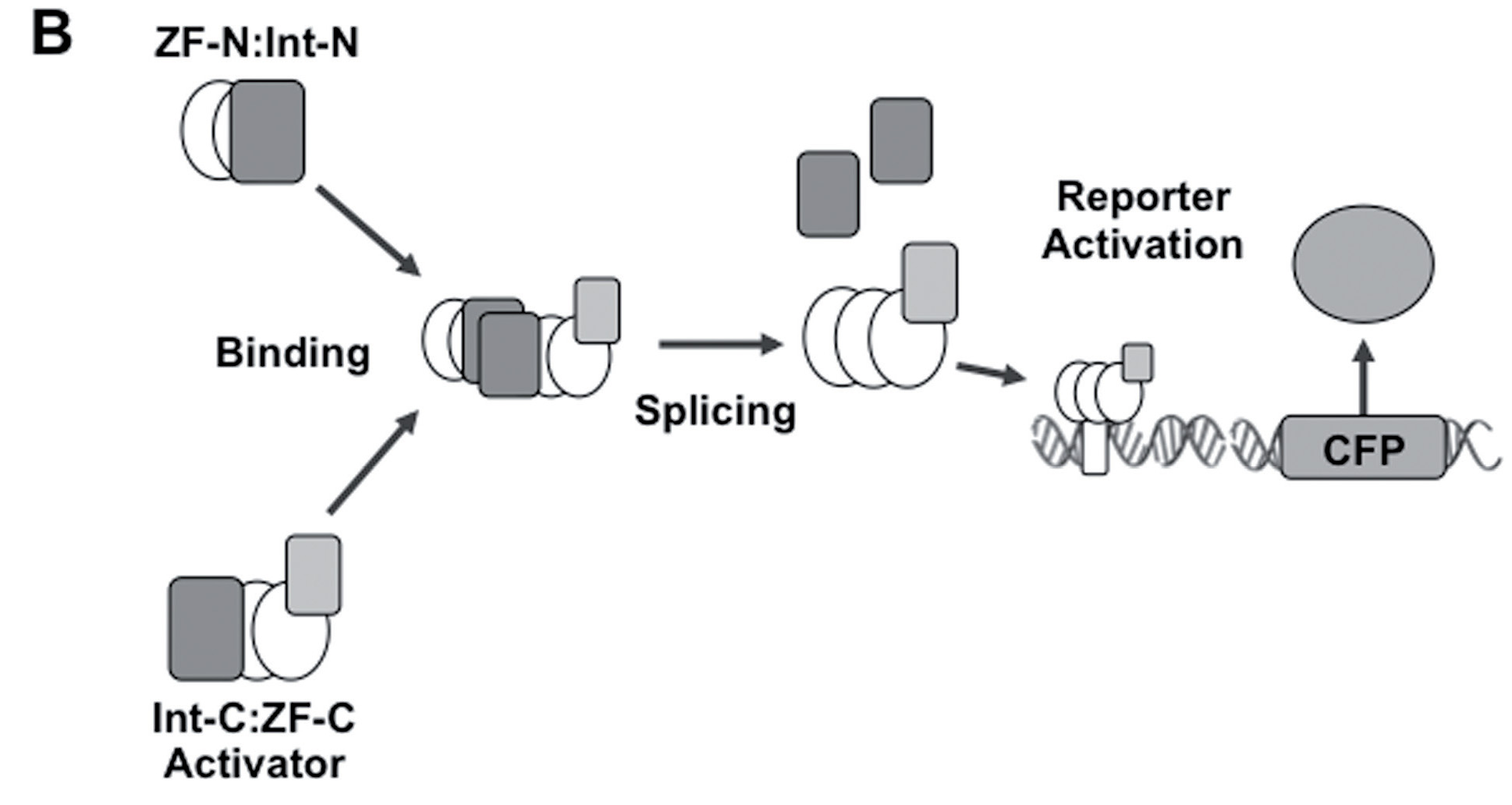
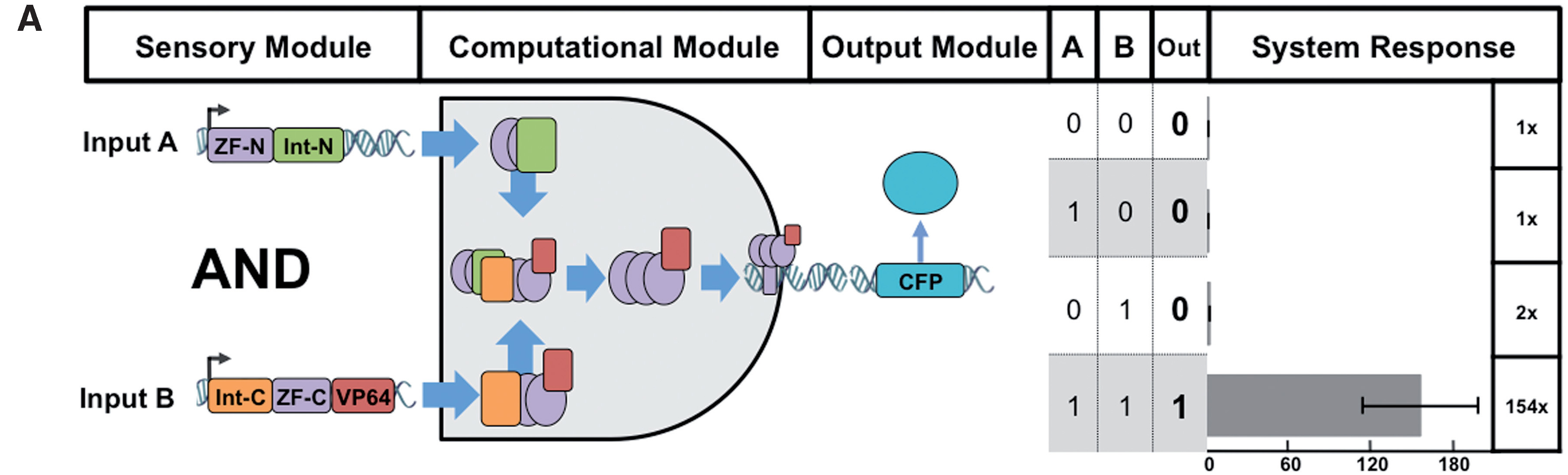
Boolean logic gates via split zinc finger-based transcription factors
Lohmueller JJ et al have demonstrated the split ZF-TF reconstitution process. Please note that we used Cfa split intein (Part:BBa_K2549009 and Part:BBa_K2549010) but not dnaB reported below.

References
- ↑ A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Lohmueller JJ, Armel TZ, Silver PA. Nucleic Acids Res, 2012 Jun;40(11):5180-7 PMID: 22323524; DOI: 10.1093/nar/gks142