Difference between revisions of "Part:BBa K2669002"
Elinramstrom (Talk | contribs) |
|||
| (13 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2669002 short</partinfo> | <partinfo>BBa_K2669002 short</partinfo> | ||
| − | BBa_K2669002 is a codon optimized AmilCP original part | + | BBa_K2669002 is a codon optimized AmilCP original native part [[Part:BBa_K592009]]. This sequence is codon optimized with the IDT optimization tool, and was optimized for E. coli K12. The native blue chromoprotein AmilCP is derived from the coral Acropora milleporais. The chromoprotein naturally exhibits a strong blue color when expressed. The color can be observed in both LB and agar culture and is visible with the naked eye. This part can be used as an indicator or as a reporter for various kinds of biological research. |
<html> | <html> | ||
| − | <img src="https://static.igem.org/mediawiki/ | + | <img src="https://static.igem.org/mediawiki/2018/9/91/T--Uppsala--Transformation_IDT_IDTbb.jpeg" width="100%" height="100%"> |
<p><b>Figure 1:</b>Transformation plate of the IDT optimized AmilCP sequence. </p> | <p><b>Figure 1:</b>Transformation plate of the IDT optimized AmilCP sequence. </p> | ||
| + | </html> | ||
| + | |||
| + | ==Contribution: Guelph 2019 == | ||
| + | |||
| + | <br> | ||
| + | This year iGEM Guelph was working with BBa_K2669002 as a reporter protein for our tetracycline biosensor. When working with this chromoprotein in <i>E.coli</i> BL21(DE3) under the control of our tetracycline operator [[BBa_K3189001]], we found that stronger colour was produced at lower temperatures compared to higher temperatures. We carried out this experiment by creating an overnight that had 5uL of it spot plated onto LB-Amp-Tet plates. The two plates were then grown overnight at 25°C and 37°C. In Figure 2, the pigment intensity was compared between two temperatures, 25°C and 37°C. It is clear from these images that at 25°C, the colonies have a darker colour compared to colonies grown at 37°C. From these results, we have characterized the expression of [[BBa_K1349002]] and can say that at 24 hours at 25°C it produces a stronger visual colour change when compared to colonies grown at 37°C for the same amount of time. This is most likely a result of reduced expression or stability of AmilCP at higher temperatures. This seemingly thermal instability makes sense given the fact that this gene was originally from coral which grows at lower temperatures in the ocean. At lower temperatures, if AmilCP is more stable, it builds up in the cells longer before degradation, resulting in darker pigmentation of the colonies. | ||
| + | <br> | ||
| + | <br> | ||
| + | https://2019.igem.org/wiki/images/thumb/f/f5/T--Guelph--AmilCPtemexpress.jpg/800px-T--Guelph--AmilCPtemexpress.jpg | ||
| + | <br> | ||
| + | <b>Figure 2: BBa_K2669002 expression at 25°C and 37°C.</b> Expression was carried out in <i>E.coli</i> BL21(DE3) on LB agar supplemented with 100 ug/mL ampicillin, 100 mg/mL tryptophan, 1mM IPTG, and 50 ng/mL tetracycline. | ||
| + | <br> | ||
Latest revision as of 03:41, 22 October 2019
AmilCP: Codon Optimized basic part
BBa_K2669002 is a codon optimized AmilCP original native part Part:BBa_K592009. This sequence is codon optimized with the IDT optimization tool, and was optimized for E. coli K12. The native blue chromoprotein AmilCP is derived from the coral Acropora milleporais. The chromoprotein naturally exhibits a strong blue color when expressed. The color can be observed in both LB and agar culture and is visible with the naked eye. This part can be used as an indicator or as a reporter for various kinds of biological research.

Figure 1:Transformation plate of the IDT optimized AmilCP sequence.
Contribution: Guelph 2019
This year iGEM Guelph was working with BBa_K2669002 as a reporter protein for our tetracycline biosensor. When working with this chromoprotein in E.coli BL21(DE3) under the control of our tetracycline operator BBa_K3189001, we found that stronger colour was produced at lower temperatures compared to higher temperatures. We carried out this experiment by creating an overnight that had 5uL of it spot plated onto LB-Amp-Tet plates. The two plates were then grown overnight at 25°C and 37°C. In Figure 2, the pigment intensity was compared between two temperatures, 25°C and 37°C. It is clear from these images that at 25°C, the colonies have a darker colour compared to colonies grown at 37°C. From these results, we have characterized the expression of BBa_K1349002 and can say that at 24 hours at 25°C it produces a stronger visual colour change when compared to colonies grown at 37°C for the same amount of time. This is most likely a result of reduced expression or stability of AmilCP at higher temperatures. This seemingly thermal instability makes sense given the fact that this gene was originally from coral which grows at lower temperatures in the ocean. At lower temperatures, if AmilCP is more stable, it builds up in the cells longer before degradation, resulting in darker pigmentation of the colonies.
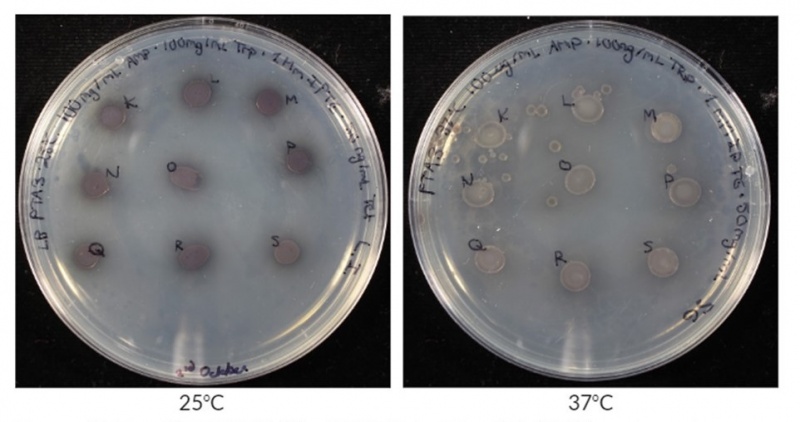
Figure 2: BBa_K2669002 expression at 25°C and 37°C. Expression was carried out in E.coli BL21(DE3) on LB agar supplemented with 100 ug/mL ampicillin, 100 mg/mL tryptophan, 1mM IPTG, and 50 ng/mL tetracycline.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
