Difference between revisions of "Part:pSB4K5"
Josephineum (Talk | contribs) |
|||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>pSB4K5 short</partinfo> | <partinfo>pSB4K5 short</partinfo> | ||
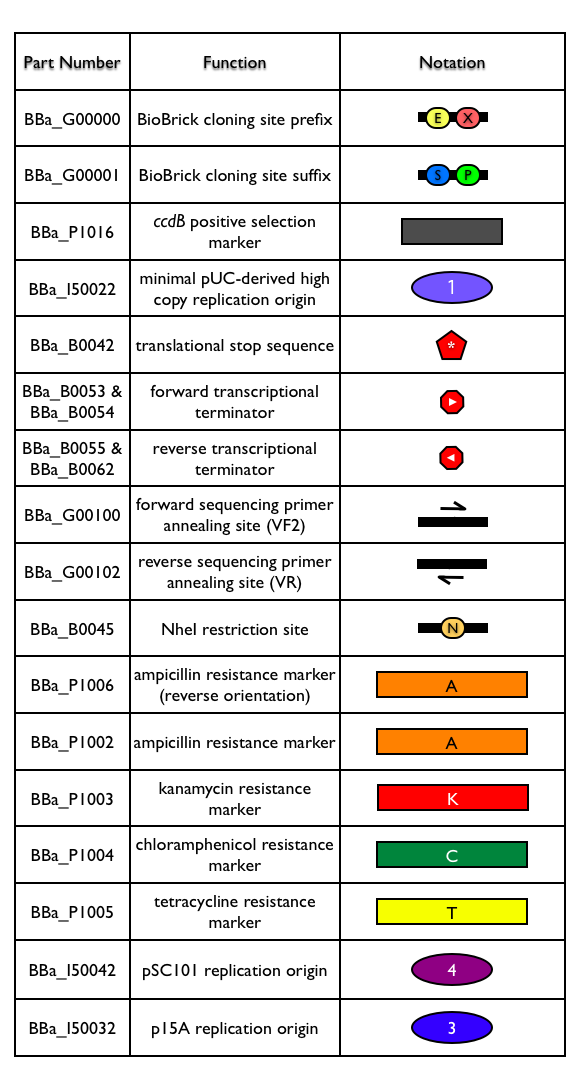
[[Part:pSB4K5|pSB4K5]] is a BioBrick standard vector with low copy pSC101 replication origin ([[Part:BBa_I50042|BBa_I50042]]) and kanamycin antibiotic resistance marker ([[Part:BBa_P1003|BBa_P1003]]). | [[Part:pSB4K5|pSB4K5]] is a BioBrick standard vector with low copy pSC101 replication origin ([[Part:BBa_I50042|BBa_I50042]]) and kanamycin antibiotic resistance marker ([[Part:BBa_P1003|BBa_P1003]]). | ||
| + | |||
| + | ===Usage and Biology=== | ||
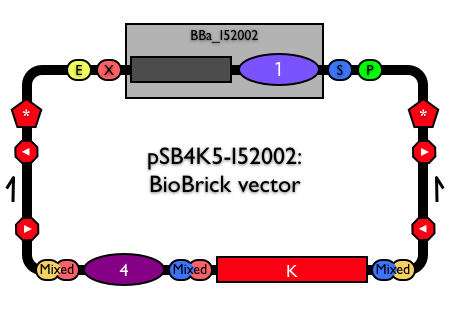
| + | [[Part:pSB4K5|pSB4K5]] is available as [[Part:pSB4K5|pSB4K5]]-[[Part:BBa_I52002|I52002]]. See [https://parts.igem.org/cgi/partsdb/dna.cgi?part_name=BBa_I52002 the physical DNA for BBa_I52002]. | ||
| + | |||
| + | ===References=== | ||
| + | '''Engineering BioBrick vectors from BioBrick parts'''<br> | ||
| + | ''Journal of Biological Engineering,'' 2008 Apr 14;2:5<br> | ||
| + | Reshma Shetty, Drew Endy, Tom Knight<br> | ||
| + | [http://www.jbioleng.org/content/2/1/5 URL]<br> | ||
| + | [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nuccore&id=169921151 GenBank EU496099] | ||
{| | {| | ||
| Line 8: | Line 19: | ||
| [[Image:BBa I51020allvectorparts.png|450px]] | | [[Image:BBa I51020allvectorparts.png|450px]] | ||
|} | |} | ||
| + | |||
| + | |||
| + | |||
| + | <h1><b>Team KCL_UK 2019</b></h1> | ||
| + | |||
| + | |||
| + | <p> | ||
| + | Our team has used this pSB4K5 part to create two plasmid fluorescent reporter system. One plasmid containing part pSB1C3 and another plasmid containing pSB4K5. This part contains part BBa_I50042 which is a pSC101 replication origin and is compatible with the minimal pUC-derived high copy replication origin found in part BBa_I50022 composite part pSB1C3 and thus suitable for E.coli transformation simultaneously with two plasmids. | ||
| + | We cloned BBa_K608010 and BBa_K608011 into this pSB4K5 plasmid backbone and created two new plasmids pSB4K5_ BBa_K608010 and pSB4K5_ BBa_K608011 respectively. Subsequently, we transformed Xl1Blue E.coli cells with these plasmids and selected positive colonies on LB agar plates containing Kanamycin (15 ug/ml). Single colony from each plate was inoculated into 10 ml LB media containing Kanamycin (15 ug/ml) and incubated overnight in a shaking incubator at 37 oC with shaking 200 rpm. After approximately 16 h of incubation E.coli cultures were diluted 1/10 with fresh 10 ml LB media containing Kanamycin (15 ug/ml) in 20 ml universal bottle. Each experiment was performed in duplicate. At this point 500 ul of the culture was collected into 1.5 ml centrifuge tube, labelled 0h incubation and stored on ice. The rest of the culture was incubated for 5 h in the shaking incubator at 37 oC with shaking 200 rpm with 500 ul samples taken every hour, labelled 1, 2, 3, 4 and 5 h incubation and stored on ice. In the end 200 ul of each duplicate sample was aliquoted into black clear bottom 96 well plate. Duplicate samples of LB media containing Kanamycin (15 ug/ml) were used as a negative control. The OD600 and fluorescence (ex485, em520) were recorded using PHERAstar FS (BMG Labtech) 96well plate reader. Recorded results were normalised to LB media and average results of two duplicate samples are presented in tables 1 and 2 and figures A and B. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | Table 1. OD600 of Xl1Blue E.coli cell culture harbouring pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 plasmids respectively. | ||
| + | </p> | ||
| + | https://2019.igem.org/wiki/images/f/f1/T--KCL_UK--characterisation_1.png | ||
| + | <p> | ||
| + | Table 2. GFP Fluorescence (ex485 nm, em520 nm) of Xl1Blue E.coli cell culture harbouring pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 plasmids respectively. | ||
| + | </p> | ||
| + | https://2019.igem.org/wiki/images/6/61/T--KCL_UK--characterisation_2.png | ||
| + | <p> | ||
| + | Figure 1. OD600 of Xl1Blue E.coli cell culture harbouring pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 plasmids respectively. | ||
| + | </p> | ||
| + | https://2019.igem.org/wiki/images/f/fe/T--KCL_UK--characterisation_3.png | ||
| + | <p> | ||
| + | Figure 2. GFP Fluorescence (ex485 nm, em520 nm) of Xl1Blue E.coli cell culture harbouring pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 plasmids respectively. | ||
| + | </p> | ||
| + | https://2019.igem.org/wiki/images/2/26/T--KCL_UK--characterisation_4.png | ||
| + | |||
| + | We also compared optical density (Table 3 and Figure 3) and fluorescence data (Table 4 and Figure 4) for pSB1C3 and pSB4K5 constructs and to our surprise we noticed a high fluorescence reading and that the replication origin of the pSB4K5 plasmid backbone might not confer a low copy number plasmid as originally thought. It has been observed that the difference in the fluorescent intensity between cells harbouring pSB1C3 and pSB4K5 plasmid GFP constructs is about 1.5-2 which might indicate that there are about 250 copies of pSB4K5 plasmid per cell not ~5 copies as in the wild-type pSC101 plasmid. | ||
| + | Similar results have been noticed by the Team Uppsala University 2012 and Team Warwick 2015 https://parts.igem.org/Part:pSB4K5:Experience. It is also known that to create part BBa_I50042 and subsequently pSB4K5 plasmids the pSC101 origin of replication had been modified to remove SpeI restriction site in the Ori region https://parts.igem.org/Part:BBa_I50042:Design, but it is not known how this modification might have changed the plasmid copy number. It has been suggested recently in the literature https://www.nature.com/articles/s41598-018-20016- wthat certain mutations in the RepA protein of the pSC101 origin of replication can increase plasmid copy number from 5 copies per cell to 113 copies per cell. Our analysis of the DNA sequence of the pSB4K5 plasmid construct has revealed two new mutations in the RepA protein R267H and H289Y but it is not clear how these changes might have influenced the differences in the plasmid copy numbers as these mutations have not been reported to date. Although differences in the plasmid copy numbers did not in any way negatively impacted on our project but it might be important for other teams to consider another plasmid backbone instead of the pSB4K5 if the low copy number plasmid is needed. | ||
| + | <p> | ||
| + | Table 3. OD600 of Xl1Blue E.coli cell culture harbouring pSB1C3_BBa_K608010, pSB1C3_BBa_K608011, pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 plasmids respectively. | ||
| + | </p> | ||
| + | https://2019.igem.org/wiki/images/e/ec/T--KCL_UK--characterisation_5.png | ||
| + | <p> | ||
| + | Figure 3. OD600 of Xl1Blue E.coli cell culture harbouring pSB1C3_BBa_K608010, pSB1C3_BBa_K608011, pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 | ||
| + | </p> | ||
| + | https://2019.igem.org/wiki/images/0/0e/T--KCL_UK--characterisation_6.png | ||
| + | <p> | ||
| + | Table 4. GFP Fluorescence (ex485 nm, em520 nm) of Xl1Blue E.coli cell culture harbouring pSB1C3_BBa_K608010, pSB1C3_BBa_K608011, pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 plasmids respectively. | ||
| + | </p> | ||
| + | https://2019.igem.org/wiki/images/1/1d/T--KCL_UK--characterisation_7.png | ||
| + | <p> | ||
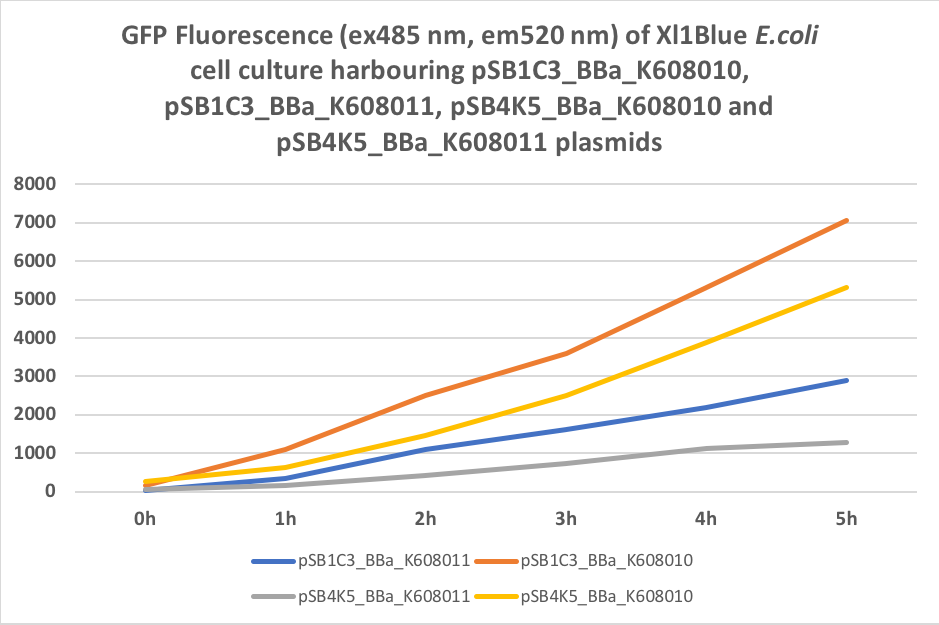
| + | Figure 4. GFP Fluorescence (ex485 nm, em520 nm) of Xl1Blue E.coli cell culture harbouring pSB1C3_BBa_K608010, pSB1C3_BBa_K608011, pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 plasmids | ||
| + | </p> | ||
| + | https://2019.igem.org/wiki/images/f/f0/T--KCL_UK--characterisation_8.png | ||
Latest revision as of 10:21, 19 October 2019
Low copy BioBrick standard vector
pSB4K5 is a BioBrick standard vector with low copy pSC101 replication origin (BBa_I50042) and kanamycin antibiotic resistance marker (BBa_P1003).
Usage and Biology
pSB4K5 is available as pSB4K5-I52002. See the physical DNA for BBa_I52002.
References
Engineering BioBrick vectors from BioBrick parts
Journal of Biological Engineering, 2008 Apr 14;2:5
Reshma Shetty, Drew Endy, Tom Knight
[http://www.jbioleng.org/content/2/1/5 URL]
[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nuccore&id=169921151 GenBank EU496099]
|
|
Team KCL_UK 2019
Our team has used this pSB4K5 part to create two plasmid fluorescent reporter system. One plasmid containing part pSB1C3 and another plasmid containing pSB4K5. This part contains part BBa_I50042 which is a pSC101 replication origin and is compatible with the minimal pUC-derived high copy replication origin found in part BBa_I50022 composite part pSB1C3 and thus suitable for E.coli transformation simultaneously with two plasmids. We cloned BBa_K608010 and BBa_K608011 into this pSB4K5 plasmid backbone and created two new plasmids pSB4K5_ BBa_K608010 and pSB4K5_ BBa_K608011 respectively. Subsequently, we transformed Xl1Blue E.coli cells with these plasmids and selected positive colonies on LB agar plates containing Kanamycin (15 ug/ml). Single colony from each plate was inoculated into 10 ml LB media containing Kanamycin (15 ug/ml) and incubated overnight in a shaking incubator at 37 oC with shaking 200 rpm. After approximately 16 h of incubation E.coli cultures were diluted 1/10 with fresh 10 ml LB media containing Kanamycin (15 ug/ml) in 20 ml universal bottle. Each experiment was performed in duplicate. At this point 500 ul of the culture was collected into 1.5 ml centrifuge tube, labelled 0h incubation and stored on ice. The rest of the culture was incubated for 5 h in the shaking incubator at 37 oC with shaking 200 rpm with 500 ul samples taken every hour, labelled 1, 2, 3, 4 and 5 h incubation and stored on ice. In the end 200 ul of each duplicate sample was aliquoted into black clear bottom 96 well plate. Duplicate samples of LB media containing Kanamycin (15 ug/ml) were used as a negative control. The OD600 and fluorescence (ex485, em520) were recorded using PHERAstar FS (BMG Labtech) 96well plate reader. Recorded results were normalised to LB media and average results of two duplicate samples are presented in tables 1 and 2 and figures A and B.
Table 1. OD600 of Xl1Blue E.coli cell culture harbouring pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 plasmids respectively.
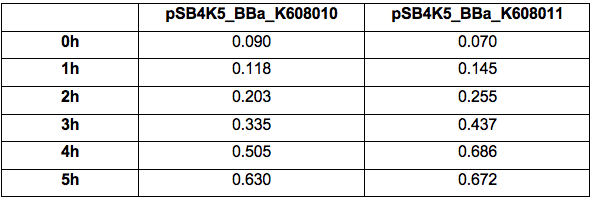
Table 2. GFP Fluorescence (ex485 nm, em520 nm) of Xl1Blue E.coli cell culture harbouring pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 plasmids respectively.
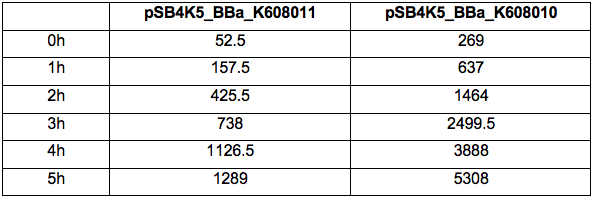
Figure 1. OD600 of Xl1Blue E.coli cell culture harbouring pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 plasmids respectively.

Figure 2. GFP Fluorescence (ex485 nm, em520 nm) of Xl1Blue E.coli cell culture harbouring pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 plasmids respectively.
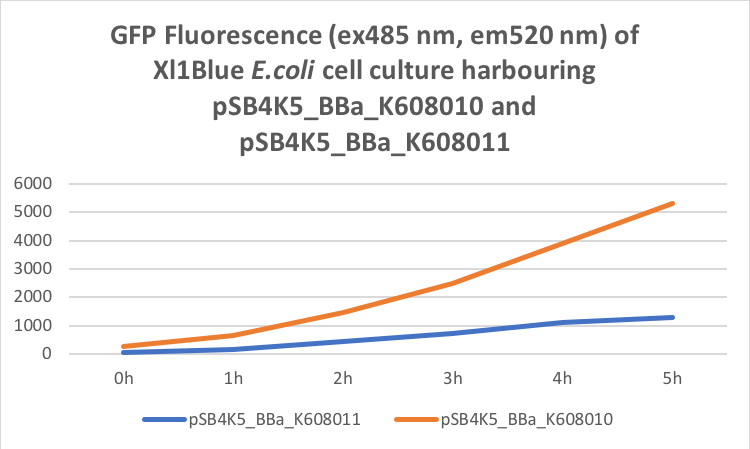
We also compared optical density (Table 3 and Figure 3) and fluorescence data (Table 4 and Figure 4) for pSB1C3 and pSB4K5 constructs and to our surprise we noticed a high fluorescence reading and that the replication origin of the pSB4K5 plasmid backbone might not confer a low copy number plasmid as originally thought. It has been observed that the difference in the fluorescent intensity between cells harbouring pSB1C3 and pSB4K5 plasmid GFP constructs is about 1.5-2 which might indicate that there are about 250 copies of pSB4K5 plasmid per cell not ~5 copies as in the wild-type pSC101 plasmid. Similar results have been noticed by the Team Uppsala University 2012 and Team Warwick 2015 https://parts.igem.org/Part:pSB4K5:Experience. It is also known that to create part BBa_I50042 and subsequently pSB4K5 plasmids the pSC101 origin of replication had been modified to remove SpeI restriction site in the Ori region https://parts.igem.org/Part:BBa_I50042:Design, but it is not known how this modification might have changed the plasmid copy number. It has been suggested recently in the literature https://www.nature.com/articles/s41598-018-20016- wthat certain mutations in the RepA protein of the pSC101 origin of replication can increase plasmid copy number from 5 copies per cell to 113 copies per cell. Our analysis of the DNA sequence of the pSB4K5 plasmid construct has revealed two new mutations in the RepA protein R267H and H289Y but it is not clear how these changes might have influenced the differences in the plasmid copy numbers as these mutations have not been reported to date. Although differences in the plasmid copy numbers did not in any way negatively impacted on our project but it might be important for other teams to consider another plasmid backbone instead of the pSB4K5 if the low copy number plasmid is needed.
Table 3. OD600 of Xl1Blue E.coli cell culture harbouring pSB1C3_BBa_K608010, pSB1C3_BBa_K608011, pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 plasmids respectively.
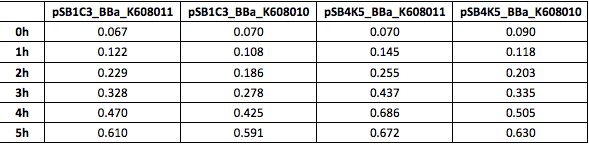
Figure 3. OD600 of Xl1Blue E.coli cell culture harbouring pSB1C3_BBa_K608010, pSB1C3_BBa_K608011, pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011
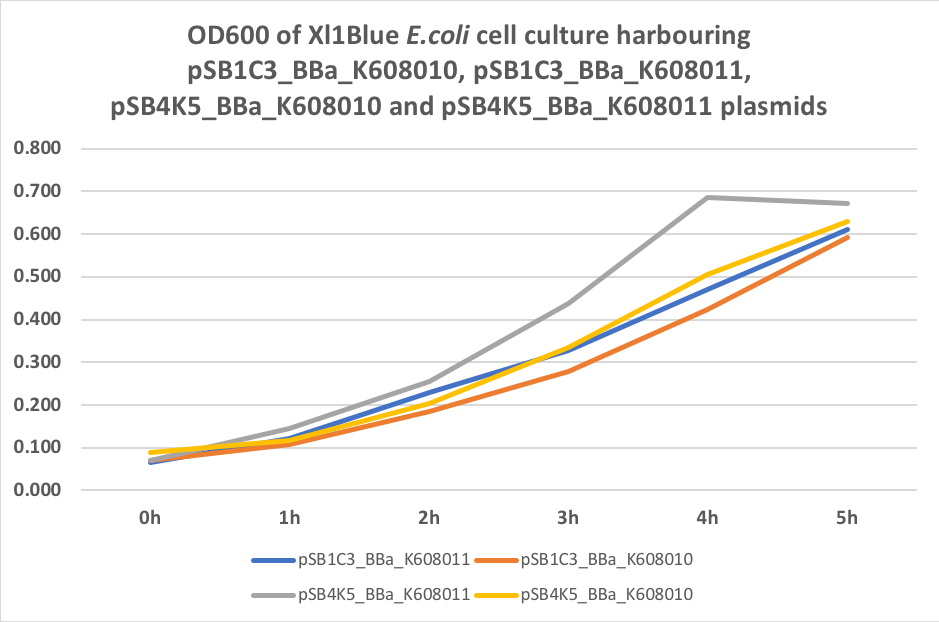
Table 4. GFP Fluorescence (ex485 nm, em520 nm) of Xl1Blue E.coli cell culture harbouring pSB1C3_BBa_K608010, pSB1C3_BBa_K608011, pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 plasmids respectively.
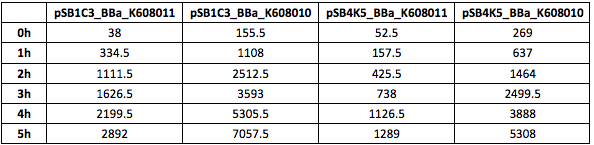
Figure 4. GFP Fluorescence (ex485 nm, em520 nm) of Xl1Blue E.coli cell culture harbouring pSB1C3_BBa_K608010, pSB1C3_BBa_K608011, pSB4K5_BBa_K608010 and pSB4K5_BBa_K608011 plasmids