Difference between revisions of "Part:BBa K2243012"
Excuseme-dzq (Talk | contribs) |
|||
| (60 intermediate revisions by 6 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K2243012 short</partinfo> | <partinfo>BBa_K2243012 short</partinfo> | ||
| + | <h2>Improvement</h2> | ||
| + | This part was improved from Part:BBa_K907000. | ||
| + | |||
| + | https://parts.igem.org/Part:BBa_K907000 | ||
| + | |||
| + | This part is improved to Part:BBa_K2557001 | ||
| + | |||
| + | 2018 NAU-CHINA obtained the sequence of this part and requested the plasmid from them. Since our chassis organism is mammalian cell, we optimized the codon and then we identified BBa_K2557001 in detail in HEK 293T cells. Although Bxb1 works well in HEK 293T cells, we did not repeat the activity reported by Peking iGEM 2017 team in E. coli. | ||
| + | |||
| + | This part is improved to Part:BBa_K3202041 | ||
| + | [http://2019.igem.org/Team:BHSF_ND BHSF_ND 2019] | ||
| + | Improved the RBS of this sequence to achieve a sharper turn for the promoter in addressing to the one on the integrase site it is acting on. Check results on [https://parts.igem.org/Part:BBa_K3202041] or down below at the bottom of the page | ||
| + | |||
| + | ===Bxb1 works well in HEK 293T cells=== | ||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/a/a9/T--NAU-CHINA-liushixibao.jpg"width="600"/> | ||
| + | </html> | ||
| + | |||
| + | Fig.2 Function verification of reversal efficiency and threshold characteristics of different recombinase in HEK 293 T Cells | ||
| + | (A)Fluorescence microscope observation of HEK 293T undergone different experimental treatments | ||
| + | (B)The statistical chart of average fluorescence intensity of cells shows that the cells with Bxb1 recombinase have a higher fluorescence intensity than those with TP901 recombinase under the same promoter strength and recombinase concentration. However, if the concentration of recombinase is low, there is no significant difference in fluorescence intensity. | ||
| + | (C)The statistics of the proportion of fluorescent cells show that the proportion of fluorescent cells has a sudden jump discontinuity between low concentration and high concentration of Bxb1 and TP901 recombinases. Similar results were obtained in all three repetitions. | ||
| + | |||
| + | |||
| + | The results of image B show that the reverse efficiency of Bxb1 recombinase is higher than TP901 recombinase under the same promoter strength and recombinase concentration. However, if the concentration of recombinase is low, there is no significant difference in fluorescence intensity. The results of the image C show that Bxb1 and TP901 recombinases have a threshold property. So, the proportion of fluorescent cells have a jump discontinuity between low concentration and high concentration of recombinase. | ||
| + | |||
| + | ===We did not repeat the activity reported by Peking iGEM 2017 team in E. coli.=== | ||
| + | Pronuclear verification of recombinase | ||
| + | |||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/3/3e/T--NAU-CHINA-junP.jpg"width="600"/> | ||
| + | </html> | ||
| + | |||
| + | Fig. 3 A total of 9 single colonies were verified. Lane 1-9, recombinase expression plasmid validation; line 10, DL2000 DNA Marker; line 11-19, reporter gene expression plasmid validation;line 20, DL2000 DNA Marker. | ||
| + | |||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/7/77/T--NAU-China--mountain1.jpg"width="200"/> | ||
| + | </html> | ||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/5/51/T--NAU-China--mountain2.jpg"width="200"/> | ||
| + | </html> | ||
| + | |||
| + | Fig. 4 Two repetitions were selected and the results showed no obvious green fluorescence | ||
| + | |||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/a/a5/T--NAU-China--mountain3.jpg"width="200"/> | ||
| + | </html> | ||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/4/49/T--NAU-CHINA-mou_4.jpg"width="200"/> | ||
| + | </html> | ||
| + | |||
| + | Fig.5 Two replicates were selected after addition of the inducer and the results showed no obvious red fluorescence | ||
| + | The result shows no obvious fluorescence. We changed some conditions, such as lowering the temperature, adjusting the rotation speed, adjusting the time, etc. But we still did not get the expected results. We consulted the teacher and the teacher replied that there might be weak fluorescence but our instrument couldn't detect it. | ||
| + | |||
| + | <h2>Abstract</h2> | ||
Bxb1 gp35 integrase comes from Mycobacterium Phage Bxb1 and can bind to specific attB/P sites to catalyze DNA recombination. It helps the phage to integrate its genome into bacterial genome naturally. | Bxb1 gp35 integrase comes from Mycobacterium Phage Bxb1 and can bind to specific attB/P sites to catalyze DNA recombination. It helps the phage to integrate its genome into bacterial genome naturally. | ||
By constructing the attB/P sites in different directions, Bxb1 gp35 can catalyze the recombination of DNA between their sites, leading to inversion when attB/P are in opposite directions and excision when attB/P are in the same directions. Bxb1 gp35 is widely used to construct combinational logic gate and performs well in changing DNA sequence | By constructing the attB/P sites in different directions, Bxb1 gp35 can catalyze the recombination of DNA between their sites, leading to inversion when attB/P are in opposite directions and excision when attB/P are in the same directions. Bxb1 gp35 is widely used to construct combinational logic gate and performs well in changing DNA sequence | ||
| − | + | <h2>Usages</h2> | |
| − | + | ||
Many researchers have paid attention to recombinases because of their ability of changing genetic circuits. Integrase Bxb1-gp35 is one of the recombinases with outstanding performance. In existence of recombinase Bxb1 gp35, different orientation of attB and attP allows the sequence to be flipped, excised, or inserted between recognition sites, which makes it useful for gene editing. In our project, we selected Bxb1-gp35 to flip the sequence flanked by attB and attP site for Bio-Flip-Flop construction. | Many researchers have paid attention to recombinases because of their ability of changing genetic circuits. Integrase Bxb1-gp35 is one of the recombinases with outstanding performance. In existence of recombinase Bxb1 gp35, different orientation of attB and attP allows the sequence to be flipped, excised, or inserted between recognition sites, which makes it useful for gene editing. In our project, we selected Bxb1-gp35 to flip the sequence flanked by attB and attP site for Bio-Flip-Flop construction. | ||
| − | + | <h2>Biology</h2> | |
| − | Integrase Bxb1-gp35 comes from Mycobacteriophage, which allows the phage to insert its DNA into the host’s genome. We got the sequence by | + | Integrase Bxb1-gp35 comes from Mycobacteriophage, which allows the phage to insert its DNA into the host’s genome. We got the sequence by de novo synthesis. |
| − | + | <h2>Design Notes</h2> | |
| − | We got the coding sequence by de novo synthesis and introduced three point mutations to remove the BsmB1 restriction enzyme cutting sites. | + | We got the coding sequence by de novo synthesis and introduced three point mutations to remove the BsmB1 restriction enzyme cutting sites(in convenience for adding RBS by Golden Gate Assenmbly later). |
| − | + | <h2>Characterization</h2> | |
| − | Since the viability of a bio-flip-flop relies on the performance of two integrases and their corresponding excisionases. To select integrases for the bio-flip-flop, we constructed expression vectors for different recombinases and tested their performance individually. | + | Since the viability of a bio-flip-flop relies on the performance of two integrases and their corresponding excisionases. To select integrases for the bio-flip-flop, we constructed expression vectors for different recombinases and tested their performance individually. |
| − | To make sure that Bxb1 have an optimal performance. We used the standard testing system, consisting of the integrase expression plasmid and the recombination reporter plasmid ( | + | To make sure that Bxb1 have an optimal performance. We used the standard testing system, consisting of the integrase expression plasmid and the recombination reporter plasmid (https://parts.igem.org/Part:BBa_K2243010). By choosing the vector with different replication origins(a p15A origin with a pTac promoter, and a ColE1 origin with a pBAD promoter) and the RBS sequences upon the integrase, we measure the recombination efficiency under different conditions.The expression vector and reporter of a recombinase were used to co-transform E. coli Top10 and samples were prepared for testing. We picked out our optimal RBS with low leakage and high efficiency for both backbone. |
| − | Flow cytometry were used to evaluate the recombination efficiency, the expression vector and reporter of a recombinase were used to co-transform E. coli Top10 and samples were prepared for flow cytometry reading. Single colonies were picked and used to inoculate 1ml of LB media with antibiotics in a V-bottom 96-well plate. The cultures were grown at 37°C and 1000 RPM for 12h. Subsequently, an aliquot comprising 2 μL of the culture was transferred into 1ml of M9 glucose media with antibiotics and inducer (1mM IPTG or 10mM arabinose for RBS tuning, gradient concentration for transfer curve) in a V-bottom 96-well plate. The cultures were grown at 37°C and 1000 RPM for 15h. An aliquot comprising 2μL of the cul-ture was transferred into 198 μL of phosphate buffered saline (PBS) containing 2 mg/mL kanamycin in a 96-well plate. This mixture was incubated for one hour at room temperature before testing. Two lasers were used to excite GFP and RFP simultaneously. Single-cell fluorescence distribution at both emission wavelengths was recorded. | + | |
| + | <html> | ||
| + | <body> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2017/f/fc/Peking_flipflop_fig_5.svg" height="500" width="600"/> | ||
| + | |||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | Fig 1. The standard testing system used to characterize the recombinases. | ||
| + | |||
| + | |||
| + | <html> | ||
| + | <body> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2017/d/dc/Peking_flipflop_fig_6.svg" height="400" width="500"/> | ||
| + | |||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | Fig 2. Integrase expression vectors with different replication origins. The one shown on top has a relaxed ColE1 replication origin and recombinase expression is induced by arabinose via a pBad induction system. The one shown in the bottom picture has a relaxed p15A replication origin and recom-binase expression is induced by IPTG. | ||
| + | |||
| + | Flow cytometry were used to evaluate the recombination efficiency, the expression vector and reporter of a recombinase were used to co-transform E. coli Top10 and samples were prepared for flow cytometry reading. Our procedures are as follows: | ||
| + | |||
| + | 1.Single colonies were picked and used to inoculate into 1ml of LB media with antibiotics in a V-bottom 96-well plate. | ||
| + | |||
| + | 2.The cultures were grown at 37°C and 1000 RPM for 12h. Subsequently, an aliquot comprising 2 μL of the culture was transferred into 1ml of M9 glucose media with antibiotics and inducer (1mM IPTG or 10mM arabinose for RBS tuning, gradient concentration for transfer curve) in a V-bottom 96-well plate. | ||
| + | |||
| + | 3.The cultures were grown at 37°C and 1000 RPM for 15h. An aliquot comprising 2μL of the cul-ture was transferred into 198 μL of phosphate buffered saline (PBS) containing 2 mg/mL kanamycin in a 96-well plate. This mixture was incubated for one hour at room temperature before testing. Two lasers were used to excite GFP and RFP simultaneously. Single-cell fluorescence distribution at both emission wavelengths was recorded. | ||
The counted cells were gated to eliminate the population which showed no fluorescence. The remaining cells were divided into two subsets by a diagonal: RFP sub-set and GFP subset. The recombination efficiency was estimated from the proportion of the RFP subset in the total fluorescent population. | The counted cells were gated to eliminate the population which showed no fluorescence. The remaining cells were divided into two subsets by a diagonal: RFP sub-set and GFP subset. The recombination efficiency was estimated from the proportion of the RFP subset in the total fluorescent population. | ||
| − | + | <html> | |
| + | <body> | ||
| − | + | <img src="https://static.igem.org/mediawiki/2017/b/ba/Peking_flipflop_fig_7.png" height="250" width="500"/> | |
| − | + | </body> | |
| + | </html> | ||
| − | Fig | + | Fig 3.A example of gating the RFP and GFP subsets. Change of fluorescence after induction was seen. Left: no inducer. Right: 10 mM Ara for 15h. |
| − | === | + | |
| + | For the vector with ColE1 replication origin, we found proper RBS in a list of RBSs for Bxb1 gp35. B0032, and RBS of 4493 TIR depicts both low leakage and high efficiency. | ||
| + | |||
| + | <html> | ||
| + | <body> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/parts/e/e5/Peking_recombinase_bxb1-rbs.png" height="300" width="400"/> | ||
| + | |||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | Fig 4. Bxb1 gp35 recombination efficiency with a variety of calculated RBS and RBS sequences from iGEM (B0030~B0035). T.I.R = Translation Initiation Rate | ||
| + | |||
| + | ===Transfer curves=== | ||
| + | |||
| + | We evaluated the performance of Bxb1 gp35 on ColE1 expression vectors under a series of inducer concentration gradients. The results enabled us to determine the appropriate inducer concentration. | ||
| + | |||
| + | <html> | ||
| + | <body> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2017/9/93/Peking_flipflop_fig17.png" height="400" width="400"/> | ||
| + | |||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | Fig 5. Bxb1 gp35 transfer curve with selected RBS, ColE1 replication origin, pBAD promoter | ||
| + | |||
| + | <html> | ||
| + | <body> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2017/0/06/Bxb1_B0032_transfer_efficiency.png" height="400" width="400"/> | ||
| + | |||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | Fig 6. Bxb1 gp35 transfer curve with RBS B0032, ColE1 replication origin, pBAD promoter | ||
| + | |||
| + | ===Fusion Protein=== | ||
| + | Besides, we construct integrase-RDF fusion protein to recognize and bind attL and attR sequences,so the state transitions become reversible.click here to read more. | ||
| + | https://parts.igem.org/Part:BBa_K2243013 | ||
| + | |||
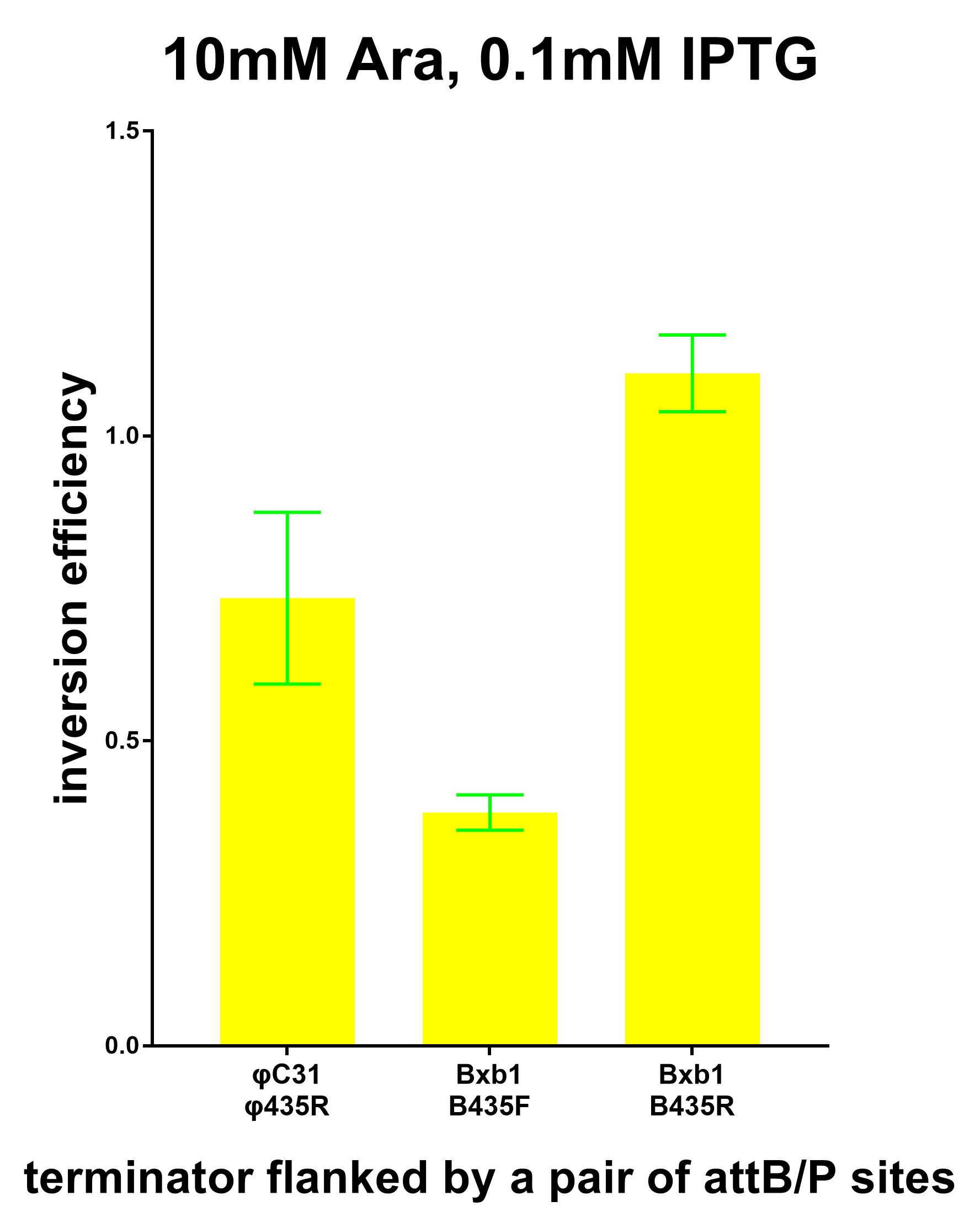
| + | ===Inversion Efficiency=== | ||
| + | Furthermore, we co-transformed the system containing the terminator flanked by Bxb1 attB/P sites and the system expressing the Bxb1 recombinase to test the inversion efficiency using the plate reader. And the results are show in the graphs below. | ||
| + | |||
| + | [[File:Peking_1pair.png|400px|thumb|center|Fig.7 Terminators flanked by one pair of attB/P sites Induced with 0.1M IPTG and 10mM Arabinose]] | ||
| + | |||
| + | [[File:Peking 3rdG inversion.png|400px|thumb|center|Fig.8 Terminators flanked by two pair of attB/P sites Induced with 0.1M IPTG and 10mM Arabinose]] | ||
| + | |||
| + | ===Enhancing the "sharpness" of the turn in the recombinase system=== | ||
| + | |||
| + | Characterised and improved by BHSF_ND 2019 | ||
| + | |||
| + | To make use of Bxb1 coding sequence from the parts collection of 2017 Peking iGEM in our recombinase flipping module, we measured the efficiency of such a recombinase system. For the project of 2017 Peking, they employed the recombinase to “turn” the state of the “clock”, “carpiod”, and “controller”, but their sequence was not particularly suited to our project. | ||
| + | |||
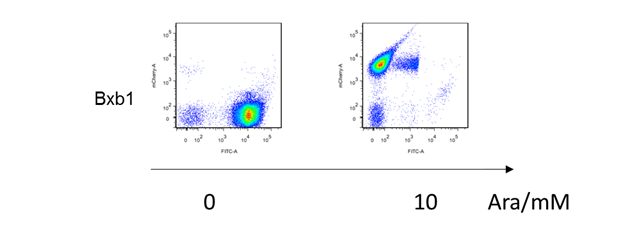
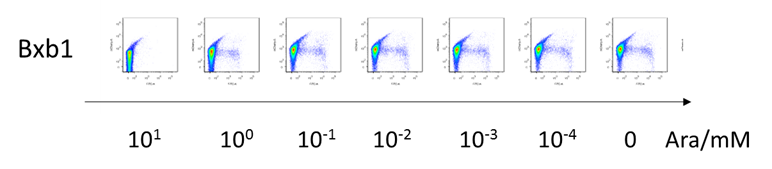
| + | [[File:T--BHSF_ND--old.png|700px|thumb|center|alt=Fluorescence pattern the recombinase|Figure 1: The x-axis of each small square represents the fluorescence of GFP, while the y-axis represents the fluorescence of RFP. Data show expression leakage in inducer present and absent. (e.g. according figures in the second row, dispersed high fluorescence of both RFP and GFP in most of the cells show that there is expression leakage of the recombinase both before and after flipping over the recombinase).]] | ||
| + | |||
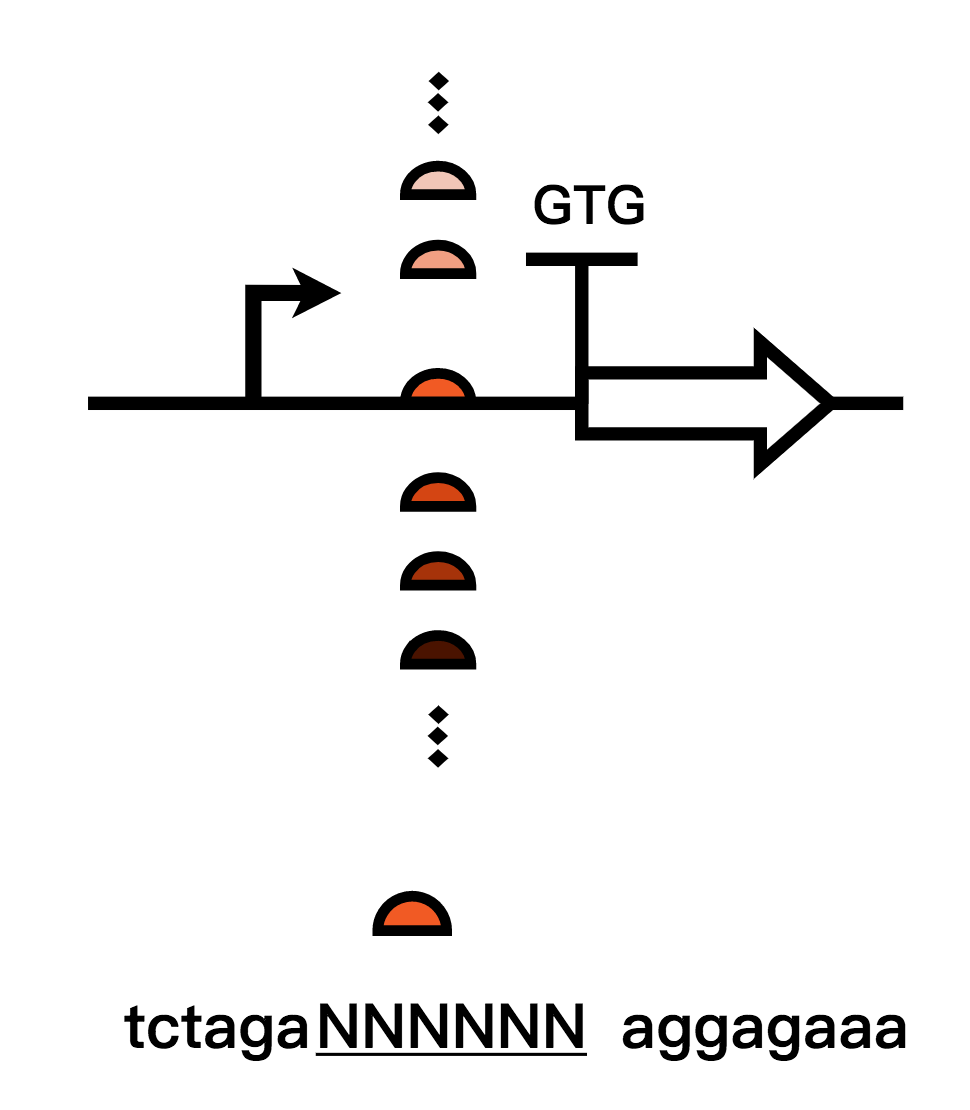
| + | We aimed to achieve “no leakage” in our system. Therefore, as for the recombinase module, a more absolute turn is needed. We turned our sight to the RBS ahead the coding sequence of Bxb1, and we tried to decrease the strength of the RBS by two commonly used methods: | ||
| + | |||
| + | 1.The first method we used is to calculated strengths of different RBS using RBS calculator and built up an RBS library to gain the weakest RBS strength and thereby reduced the expression leakage(In addressing to our project)of Bxb1; | ||
| + | |||
| + | 2. The second method is that we mutated the original RBS sequence, screened them and changed the start codon of RBS from ATG to GTG in order to reduce recombinase expression leakage levels(In addressing to our project). We chose the optimal one and retested them. | ||
| + | |||
| + | After the improvement we retested the effectiveness of the system again quantitatively by flow cytometry. Graphs shown below demonstrated little expression leakage before and after the expression of recombinase since fluorescence of most cells concentrates at either high fluorescence of GFP in inducer’s present, before flipping, or high fluorescence of RFP in inducer’s absent, after flipping. | ||
| + | |||
| + | [[File:T--BHSF ND--Recombinase design 3.png|425px|thumb|center|alt=Two means of improvement |Figure 2: two means of improvement]] | ||
| + | |||
| + | [[File:T--BHSF ND--improved.png|700px|thumb|right|alt=Improved fluorescence pattern for the recombinase|Figure 3: Improved fluorescence pattern for the recombinase]] | ||
| + | |||
| + | ====Discussion==== | ||
| + | |||
| + | Our recombinases can effectively invert the in-between promoter under our design and experimental validation. This feature works well as a reactor for changes in the external environment. It permanently changes the sequence of genes and thus the direction of gene expression, which implements our goal of connecting two bistable system and exerting permanent effect on the circuit. | ||
| + | |||
| + | We improved the expression leakage of the irreversible recombinase in order to ensure that no leaked recombinase is present in inducer’s presence which may accidentally initiates the expression of toxin. Aided with that improvement, the expression of RFP corresponds with the “0” state when applied to our circuit, where the bacterium hasn’t entered its working condition; while the expression of GFP after P1 has been reversed corresponds with the “standby” state in our circuit. | ||
| + | |||
| + | |||
| + | <h2>Source</h2> | ||
Mycobacterium Phage Bxb1 | Mycobacterium Phage Bxb1 | ||
| − | + | ||
| + | <h2>References</h2> | ||
Roquet, N., Soleimany, A.P., Ferris, A.C., Aaronson, S. & Lu, T.K. Synthetic recombinase-based state machines in living cells. Science 353, aad8559 (2016). | Roquet, N., Soleimany, A.P., Ferris, A.C., Aaronson, S. & Lu, T.K. Synthetic recombinase-based state machines in living cells. Science 353, aad8559 (2016). | ||
| + | |||
| + | |||
Latest revision as of 00:23, 22 October 2019
Bxb1 gp35
Contents
Improvement
This part was improved from Part:BBa_K907000.
https://parts.igem.org/Part:BBa_K907000
This part is improved to Part:BBa_K2557001
2018 NAU-CHINA obtained the sequence of this part and requested the plasmid from them. Since our chassis organism is mammalian cell, we optimized the codon and then we identified BBa_K2557001 in detail in HEK 293T cells. Although Bxb1 works well in HEK 293T cells, we did not repeat the activity reported by Peking iGEM 2017 team in E. coli.
This part is improved to Part:BBa_K3202041 [http://2019.igem.org/Team:BHSF_ND BHSF_ND 2019] Improved the RBS of this sequence to achieve a sharper turn for the promoter in addressing to the one on the integrase site it is acting on. Check results on [1] or down below at the bottom of the page
Bxb1 works well in HEK 293T cells

Fig.2 Function verification of reversal efficiency and threshold characteristics of different recombinase in HEK 293 T Cells (A)Fluorescence microscope observation of HEK 293T undergone different experimental treatments (B)The statistical chart of average fluorescence intensity of cells shows that the cells with Bxb1 recombinase have a higher fluorescence intensity than those with TP901 recombinase under the same promoter strength and recombinase concentration. However, if the concentration of recombinase is low, there is no significant difference in fluorescence intensity. (C)The statistics of the proportion of fluorescent cells show that the proportion of fluorescent cells has a sudden jump discontinuity between low concentration and high concentration of Bxb1 and TP901 recombinases. Similar results were obtained in all three repetitions.
The results of image B show that the reverse efficiency of Bxb1 recombinase is higher than TP901 recombinase under the same promoter strength and recombinase concentration. However, if the concentration of recombinase is low, there is no significant difference in fluorescence intensity. The results of the image C show that Bxb1 and TP901 recombinases have a threshold property. So, the proportion of fluorescent cells have a jump discontinuity between low concentration and high concentration of recombinase.
We did not repeat the activity reported by Peking iGEM 2017 team in E. coli.
Pronuclear verification of recombinase

Fig. 3 A total of 9 single colonies were verified. Lane 1-9, recombinase expression plasmid validation; line 10, DL2000 DNA Marker; line 11-19, reporter gene expression plasmid validation;line 20, DL2000 DNA Marker.


Fig. 4 Two repetitions were selected and the results showed no obvious green fluorescence


Fig.5 Two replicates were selected after addition of the inducer and the results showed no obvious red fluorescence The result shows no obvious fluorescence. We changed some conditions, such as lowering the temperature, adjusting the rotation speed, adjusting the time, etc. But we still did not get the expected results. We consulted the teacher and the teacher replied that there might be weak fluorescence but our instrument couldn't detect it.
Abstract
Bxb1 gp35 integrase comes from Mycobacterium Phage Bxb1 and can bind to specific attB/P sites to catalyze DNA recombination. It helps the phage to integrate its genome into bacterial genome naturally. By constructing the attB/P sites in different directions, Bxb1 gp35 can catalyze the recombination of DNA between their sites, leading to inversion when attB/P are in opposite directions and excision when attB/P are in the same directions. Bxb1 gp35 is widely used to construct combinational logic gate and performs well in changing DNA sequence
Usages
Many researchers have paid attention to recombinases because of their ability of changing genetic circuits. Integrase Bxb1-gp35 is one of the recombinases with outstanding performance. In existence of recombinase Bxb1 gp35, different orientation of attB and attP allows the sequence to be flipped, excised, or inserted between recognition sites, which makes it useful for gene editing. In our project, we selected Bxb1-gp35 to flip the sequence flanked by attB and attP site for Bio-Flip-Flop construction.
Biology
Integrase Bxb1-gp35 comes from Mycobacteriophage, which allows the phage to insert its DNA into the host’s genome. We got the sequence by de novo synthesis.
Design Notes
We got the coding sequence by de novo synthesis and introduced three point mutations to remove the BsmB1 restriction enzyme cutting sites(in convenience for adding RBS by Golden Gate Assenmbly later).
Characterization
Since the viability of a bio-flip-flop relies on the performance of two integrases and their corresponding excisionases. To select integrases for the bio-flip-flop, we constructed expression vectors for different recombinases and tested their performance individually.
To make sure that Bxb1 have an optimal performance. We used the standard testing system, consisting of the integrase expression plasmid and the recombination reporter plasmid (https://parts.igem.org/Part:BBa_K2243010). By choosing the vector with different replication origins(a p15A origin with a pTac promoter, and a ColE1 origin with a pBAD promoter) and the RBS sequences upon the integrase, we measure the recombination efficiency under different conditions.The expression vector and reporter of a recombinase were used to co-transform E. coli Top10 and samples were prepared for testing. We picked out our optimal RBS with low leakage and high efficiency for both backbone.

Fig 1. The standard testing system used to characterize the recombinases.

Fig 2. Integrase expression vectors with different replication origins. The one shown on top has a relaxed ColE1 replication origin and recombinase expression is induced by arabinose via a pBad induction system. The one shown in the bottom picture has a relaxed p15A replication origin and recom-binase expression is induced by IPTG.
Flow cytometry were used to evaluate the recombination efficiency, the expression vector and reporter of a recombinase were used to co-transform E. coli Top10 and samples were prepared for flow cytometry reading. Our procedures are as follows:
1.Single colonies were picked and used to inoculate into 1ml of LB media with antibiotics in a V-bottom 96-well plate.
2.The cultures were grown at 37°C and 1000 RPM for 12h. Subsequently, an aliquot comprising 2 μL of the culture was transferred into 1ml of M9 glucose media with antibiotics and inducer (1mM IPTG or 10mM arabinose for RBS tuning, gradient concentration for transfer curve) in a V-bottom 96-well plate.
3.The cultures were grown at 37°C and 1000 RPM for 15h. An aliquot comprising 2μL of the cul-ture was transferred into 198 μL of phosphate buffered saline (PBS) containing 2 mg/mL kanamycin in a 96-well plate. This mixture was incubated for one hour at room temperature before testing. Two lasers were used to excite GFP and RFP simultaneously. Single-cell fluorescence distribution at both emission wavelengths was recorded.
The counted cells were gated to eliminate the population which showed no fluorescence. The remaining cells were divided into two subsets by a diagonal: RFP sub-set and GFP subset. The recombination efficiency was estimated from the proportion of the RFP subset in the total fluorescent population.

Fig 3.A example of gating the RFP and GFP subsets. Change of fluorescence after induction was seen. Left: no inducer. Right: 10 mM Ara for 15h.
For the vector with ColE1 replication origin, we found proper RBS in a list of RBSs for Bxb1 gp35. B0032, and RBS of 4493 TIR depicts both low leakage and high efficiency.

Fig 4. Bxb1 gp35 recombination efficiency with a variety of calculated RBS and RBS sequences from iGEM (B0030~B0035). T.I.R = Translation Initiation Rate
Transfer curves
We evaluated the performance of Bxb1 gp35 on ColE1 expression vectors under a series of inducer concentration gradients. The results enabled us to determine the appropriate inducer concentration.

Fig 5. Bxb1 gp35 transfer curve with selected RBS, ColE1 replication origin, pBAD promoter

Fig 6. Bxb1 gp35 transfer curve with RBS B0032, ColE1 replication origin, pBAD promoter
Fusion Protein
Besides, we construct integrase-RDF fusion protein to recognize and bind attL and attR sequences,so the state transitions become reversible.click here to read more. https://parts.igem.org/Part:BBa_K2243013
Inversion Efficiency
Furthermore, we co-transformed the system containing the terminator flanked by Bxb1 attB/P sites and the system expressing the Bxb1 recombinase to test the inversion efficiency using the plate reader. And the results are show in the graphs below.
Enhancing the "sharpness" of the turn in the recombinase system
Characterised and improved by BHSF_ND 2019
To make use of Bxb1 coding sequence from the parts collection of 2017 Peking iGEM in our recombinase flipping module, we measured the efficiency of such a recombinase system. For the project of 2017 Peking, they employed the recombinase to “turn” the state of the “clock”, “carpiod”, and “controller”, but their sequence was not particularly suited to our project.
We aimed to achieve “no leakage” in our system. Therefore, as for the recombinase module, a more absolute turn is needed. We turned our sight to the RBS ahead the coding sequence of Bxb1, and we tried to decrease the strength of the RBS by two commonly used methods:
1.The first method we used is to calculated strengths of different RBS using RBS calculator and built up an RBS library to gain the weakest RBS strength and thereby reduced the expression leakage(In addressing to our project)of Bxb1;
2. The second method is that we mutated the original RBS sequence, screened them and changed the start codon of RBS from ATG to GTG in order to reduce recombinase expression leakage levels(In addressing to our project). We chose the optimal one and retested them.
After the improvement we retested the effectiveness of the system again quantitatively by flow cytometry. Graphs shown below demonstrated little expression leakage before and after the expression of recombinase since fluorescence of most cells concentrates at either high fluorescence of GFP in inducer’s present, before flipping, or high fluorescence of RFP in inducer’s absent, after flipping.
Discussion
Our recombinases can effectively invert the in-between promoter under our design and experimental validation. This feature works well as a reactor for changes in the external environment. It permanently changes the sequence of genes and thus the direction of gene expression, which implements our goal of connecting two bistable system and exerting permanent effect on the circuit.
We improved the expression leakage of the irreversible recombinase in order to ensure that no leaked recombinase is present in inducer’s presence which may accidentally initiates the expression of toxin. Aided with that improvement, the expression of RFP corresponds with the “0” state when applied to our circuit, where the bacterium hasn’t entered its working condition; while the expression of GFP after P1 has been reversed corresponds with the “standby” state in our circuit.
Source
Mycobacterium Phage Bxb1
References
Roquet, N., Soleimany, A.P., Ferris, A.C., Aaronson, S. & Lu, T.K. Synthetic recombinase-based state machines in living cells. Science 353, aad8559 (2016).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 192
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 466
Illegal XhoI site found at 553 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1105
Illegal NgoMIV site found at 1192
Illegal AgeI site found at 242 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1300