Part:BBa_K3202041
AraC-Pc-pBAD-Bxb1 Improved
This part was improved from Part:BBa_K2243012
This composite part is a improved version of Bxb1 recombinase coding sequence
In our study, we find out that the recombinase translation is not quite effecient. Quantitatively measured this issue, we find that the translated products have shown a indecisiveness addressing to the expression of sfGFP and mRFP for the integrase site. See figure 1. After certain modification on the RBS of the coding sequnce for the recombinase, the switch ON/OFF resulted is more absolute. See figure 2.
Recombinase Flipping Module
I.Background
We sought for a structure which could implement permanent memory on changes in our circuit while effectively connects our double bistable system and were inspired by the design of recombinase in Peking 2017. We tested three types of recombinase, Bxb1, TP901, PhiC31 they had in their project in our experiment.
Scientists discovered that phages phiC31 integrase make use of LSTP integrases in mediating phage integration and excision into the bacterial genome between their cognate recognition sites, attB (bacterium) and attP (phage). “By placing these sites in the opposite orientation, LSTP integrases cleave, rotate and rejoin the DNA to invert the region between sites. As shown in figure 1, LSTP integrases catalyze insertion of phage genome (yellow) into the bacterial genome (blue) between attB and attP sites, which form hybrid attL and attR sites. Multicolored arrowheads illustrate the sequence changes that occur during strand exchange, with the core sequence shown in yellow.”
We make use of the property of irreversible recombinase which can implement permanent memory so that before reverting the recombinase the second layer, the DNase, is inhibited in working condition, and after reverting the second layer can be eternally expressed, preventing the bacterium from reviving through other techniques.
II.Design
In our team, we include the recombinase in order to “implement the permanent memory of the circuit by irreversibly inverting the orientation of the intervening PR1 according to article Permanent genetic memory with >1-byte capacity.” The recombinase is able to flip the DNA in only one direction and thus implement permanent memory.
When it is applied to our circuit, P1 is inverted in regular working condition where inducer is present, recombinase catalyzes the cleavage of each cognate recognition sites attB and attP, inverting the orientation of P1 and rejoining the circuit at attL and attR respectively. Due to the fact that irreversible recombinase can only flip the DNA in one direction, the inverted P1 which is inhibited in working condition where inducer is present remains inverted even when the recombinase is not expressed as inducer is absent. When the inducer is absent, R1 doesn’t inhibit the expression of the inverted P1 anymore, the toxin in the second layer can be then expressed and conduct suicide. The inversion is a permanent effect on the bistable system, that is to say even if we stimulate the whole system again by adding inducer, P1 will not switch its orientation back.
III.Experiment & Results
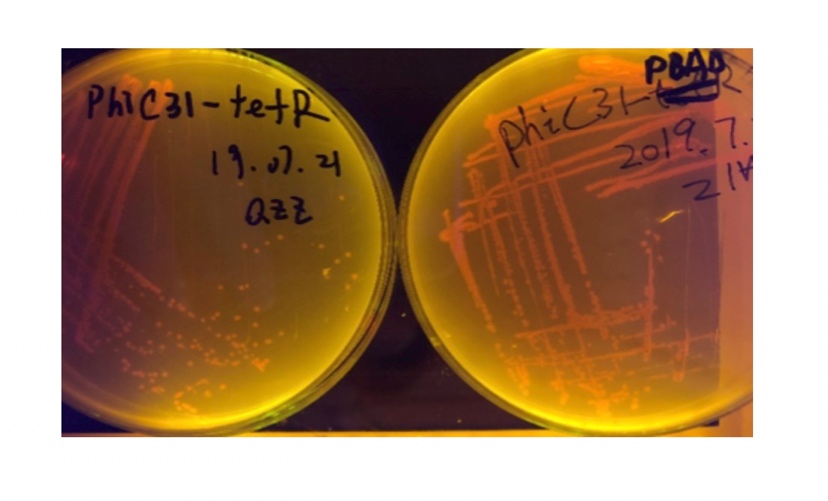
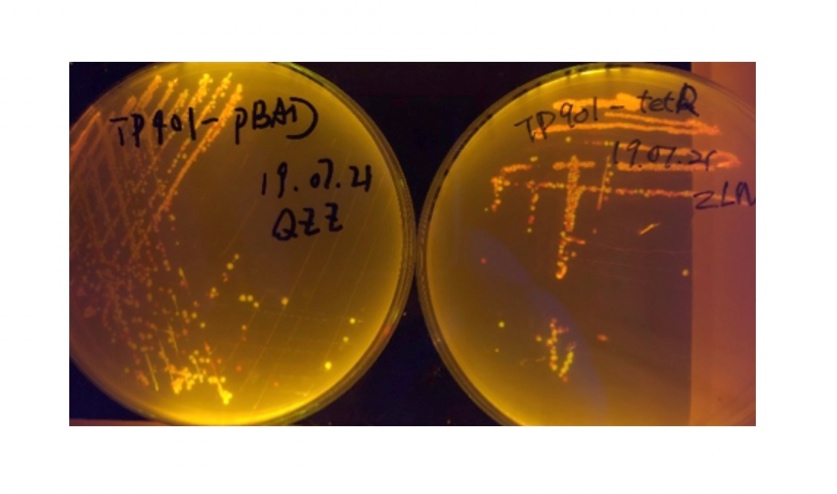
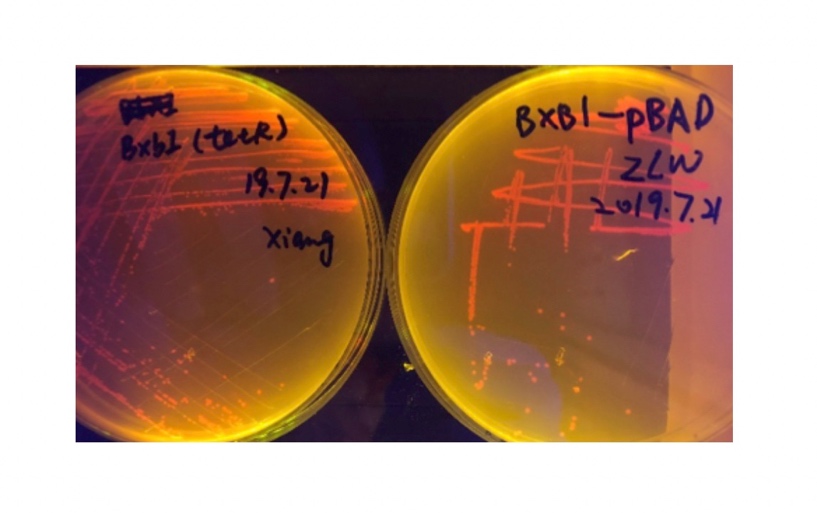
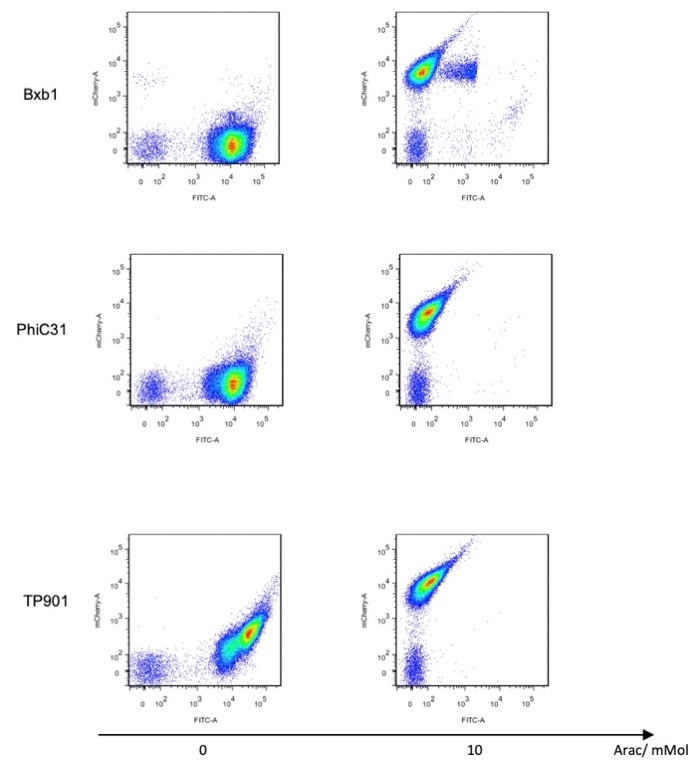
We used pBAD and PLtetO as promoters to regulate the expression of the three recombinases in order to test the effect flipping over the promoter J23119 (P1) by the three recombinases and to see if there is expression leakage.
Theoretically speaking, when inducer is present, recombinase should be expressed and successfully reverts the promoter P1 in-between, the bacterium will express mRFP, displaying red color; while if no inducer is present, the inhibitory effect the transcriptional factor exerts on P2 will stop the expression of recombinase, P1 fails to reverse, the bacterium will therefore express sfGFP, displaying green color.
In the plating medium with no inducer, the leaked expression of recombinase reverted the promoter J23119, displaying red color. Comparing the three sets of recombinases (Fig. 3&4&5), we found that TP901 has the lowest expression leakage.
Then we used pBAD to test the recombinases’ working efficiency in inducer’s presence through flow cytometry. Results are quantitatively shown in the diagram below.
Improvement
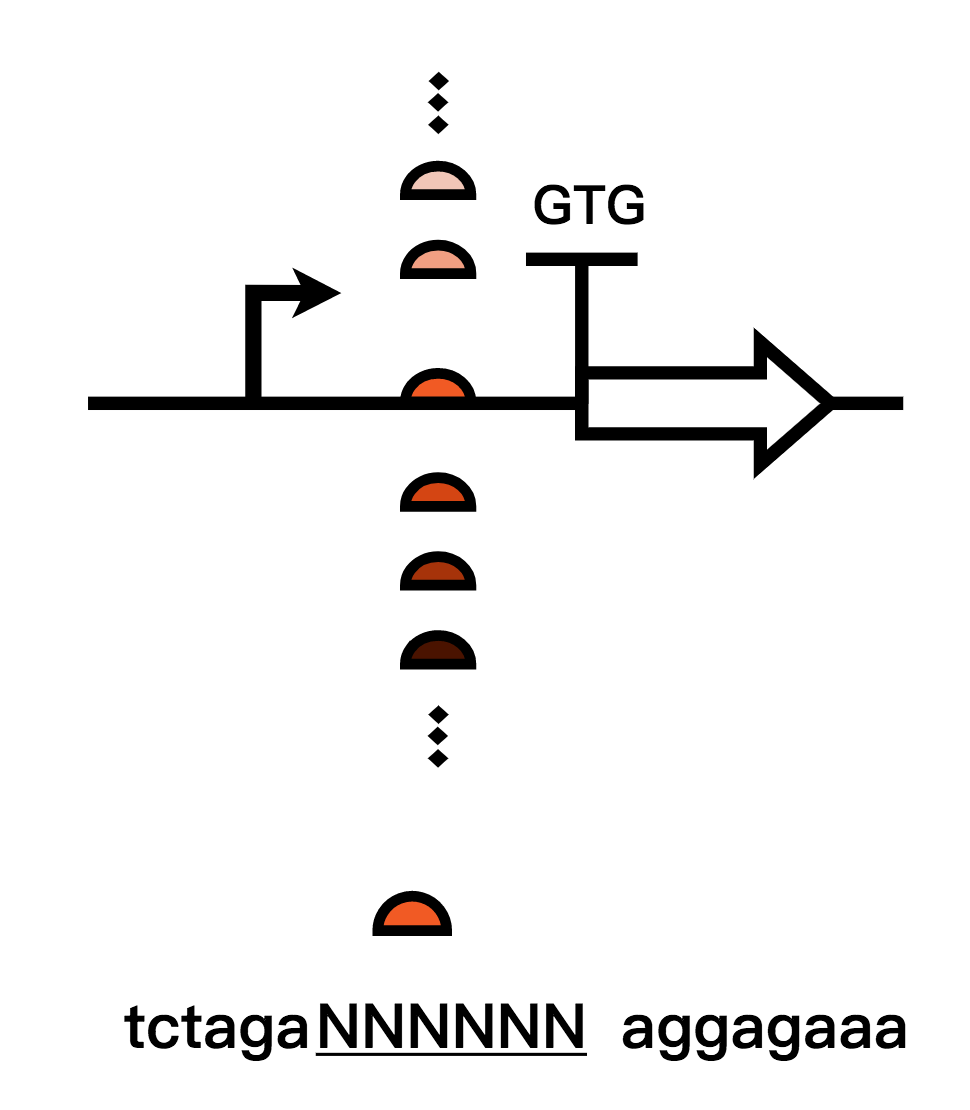
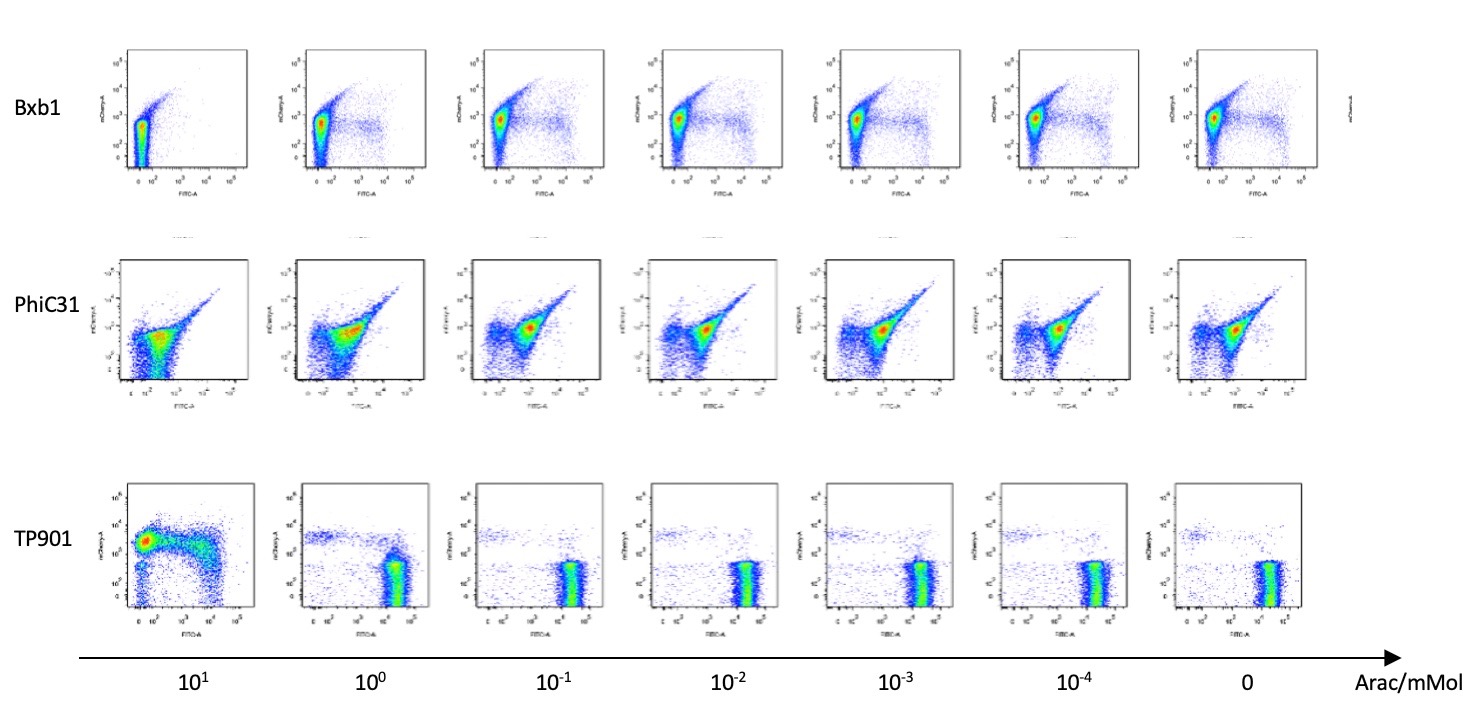
To make use of Bxb1 coding sequence from the parts collection of 2017 Peking iGEM in our recombinase flipping module, we measured the efficiency of such a recombinase system. For the project of 2017 Peking, they employed the recombinase to “turn” the state of the “clock”, “carpiod”, and “controller”, but their sequence was not particularly suited to our project. We aimed to achieve “no leakage” in our system. Therefore, as for the recombinase module, a more absolute turn is needed. We turned our sight to the RBS ahead the coding sequence of Bxb1, and we tried to decrease the strength of the RBS by two commonly used methods:
1.The first method we used is to calculated strengths of different RBS using RBS calculator and built up an RBS library to gain the weakest RBS strength and thereby reduced the expression leakage(In addressing to our project)of Bxb1;
2. The second method is that we mutated the original RBS sequence, screened them and changed the start codon of RBS from ATG to GTG in order to reduce recombinase expression leakage levels(In addressing to our project). We chose the optimal one and retested them.
After the improvement we retested the effectiveness of the system again quantitatively by flow cytometry. Graphs shown below demonstrated little expression leakage before and after the expression of recombinase since fluorescence of most cells concentrates at either high fluorescence of GFP in inducer’s present, before flipping, or high fluorescence of RFP in inducer’s absent, after flipping.
IV.Discussion
Our recombinases can effectively invert the in-between promoter under our design and experimental validation. This feature works well as a reactor for changes in the external environment. It permanently changes the sequence of genes and thus the direction of gene expression, which implements our goal of connecting two bistable system and exerting permanent effect on the circuit.
We improved the expression leakage of the irreversible recombinase in order to ensure that no leaked recombinase is present in inducer’s presence which may accidentally initiates the expression of toxin. Aided with that improvement, the expression of RFP corresponds with the “0” state when applied to our circuit, where the bacterium hasn’t entered its working condition; while the expression of GFP after P1 has been reversed corresponds with the “standby” state in our circuit.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1307
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1033
Illegal BamHI site found at 1866
Illegal XhoI site found at 946 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 307
Illegal NgoMIV site found at 394
Illegal AgeI site found at 1257
Illegal AgeI site found at 1701 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 199
Illegal SapI site found at 1683
| None |