Difference between revisions of "Chassis/Cell-Free Systems"
(→Cell-Free Systems investigated) |
Samanthapeb (Talk | contribs) m (→Advantages and disadvantages of CFS) |
||
| (22 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
=Cell-Free Systems= | =Cell-Free Systems= | ||
| + | __NOTOC__ | ||
| + | Please click on each system for its individual characterization. | ||
| + | {| border="0" width="100%" | ||
| + | |- align=center | ||
| + | ||[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30 <font face=georgia color=#330033 size=4>Commercial ''E. coli'' S30</font>] | ||
| + | ||[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_T7_S30 <font face=georgia color=#330033 size=4>Commercial ''E. coli'' T7 S30</font>] | ||
| + | ||[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Homemade_E.coli_S30 <font face=georgia color=#330033 size=4>Homemade ''E. coli'' S30</font>] | ||
| + | ||[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Vesicle <font face=georgia color=#330033 size=4>Vesicle-encapsulation</font>] | ||
| + | |- | ||
| + | |} | ||
| + | |||
==Introduction== | ==Introduction== | ||
'''Cell-Free Systems (CFS)''' involve the in-vitro expression of genes into proteins. These systems can serve as a compatible chassis for the various parts and devices from the [https://parts.igem.org/wiki/index.php/Main_Page Registry of Standard Biological Parts]. | '''Cell-Free Systems (CFS)''' involve the in-vitro expression of genes into proteins. These systems can serve as a compatible chassis for the various parts and devices from the [https://parts.igem.org/wiki/index.php/Main_Page Registry of Standard Biological Parts]. | ||
| Line 20: | Line 31: | ||
|width=50%|<center>'''Disadvantages'''</center> | |width=50%|<center>'''Disadvantages'''</center> | ||
|- | |- | ||
| − | |style="background:#eeffee"|System is | + | |style="background:#eeffee"|System is not an organism and is not restricted by the policies imposed on genetically modified organisms(GMO) |
|style="background:#ffeeee"|Short expression lifespan because of limited energy of the system even in the presence of an ATP regenerating system | |style="background:#ffeeee"|Short expression lifespan because of limited energy of the system even in the presence of an ATP regenerating system | ||
|- | |- | ||
|style="background:#eeffee"|Process is quick and simple requiring only preparation of cell extract and feeding solution and subsequent addition of DNA template | |style="background:#eeffee"|Process is quick and simple requiring only preparation of cell extract and feeding solution and subsequent addition of DNA template | ||
| − | |style="background:#ffeeee"|Expensive | + | |style="background:#ffeeee"|Expensive system has no sustained metabolism to convert cheap energy (like sugars) into useable one for the gene expression machinery |
|- | |- | ||
| − | |style="background:#eeffee"| | + | |style="background:#eeffee"|No concurrent expression of existing genome, therefore your genetically engineered device is more energy efficient |
|style="background:#ffeeee"|Less characterization and experience of use in the laboratories compared to ''E. coli'' | |style="background:#ffeeee"|Less characterization and experience of use in the laboratories compared to ''E. coli'' | ||
| + | |- | ||
| + | |style="background:#eeffee"|No DNA mutation of your genetically engineered device because there is no DNA replication | ||
| + | |style="background:#ffeeee"| | ||
| + | |- | ||
| + | |style="background:#eeffee"|No selective pressue on your genetically engineered device because the system is non-living and does not undergo natural selection | ||
| + | |style="background:#ffeeee"| | ||
| + | |- | ||
| + | |style="background:#eeffee"|No self-replication of your genetically engineered device leads to a fixed amount of DNA being expressed and more control over the rate of expression | ||
| + | |style="background:#ffeeee"| | ||
| + | |- | ||
| + | |style="background:#eeffee"|Expression system can be quality-controlled by manipulating adjustable parameters e.g. buffers are added to maintain optimum magnesium concentrations for efficient translation; protease inhibitors can be added to minimize degradation of synthesized proteins | ||
| + | |style="background:#ffeeee"| | ||
|} | |} | ||
<br> | <br> | ||
| Line 33: | Line 56: | ||
==Specifications for CFS characterization== | ==Specifications for CFS characterization== | ||
Several parameters are identified to help us understand the advantages and disadvantages associated with using a particular chassis. | Several parameters are identified to help us understand the advantages and disadvantages associated with using a particular chassis. | ||
| + | [[Image:CFS_Specifications.png|thumb|350px|right|CFS Specifications (hypothetical graph)]] | ||
{| border="1" | {| border="1" | ||
|- | |- | ||
| Line 41: | Line 65: | ||
||Ability to express Biobricks | ||Ability to express Biobricks | ||
|- | |- | ||
| − | |style="background:#ffffcc"| | + | |style="background:#ffffcc"|Temperature dependence |
| − | || | + | ||The relationship between expression rate and temperature. |
|- | |- | ||
|style="background:#ffffcc"|Optimum amount of DNA | |style="background:#ffffcc"|Optimum amount of DNA | ||
| Line 58: | Line 82: | ||
|} | |} | ||
To gain knowledge about the CFS, we have selected the following parts available in the registry to extract the above-mentioned properties. | To gain knowledge about the CFS, we have selected the following parts available in the registry to extract the above-mentioned properties. | ||
| − | *A simple constitutive gene expression device that | + | *A simple constitutive gene expression device that requires an ''E. coli'' RNA polymerase [https://parts.igem.org/wiki/index.php/Part:BBa_I13522 BBa_I13522] |
| − | *A simple constitutive gene expression device that requires a T7 bacteriophage RNA polymerase [https://parts.igem.org/wiki/index.php/Part: | + | *A simple constitutive gene expression device that requires a T7 bacteriophage RNA polymerase [https://parts.igem.org/wiki/index.php/Part:BBa_I719015 BBa_I719015] |
*An inducible gene expression device that is well-characterized in the registry [https://parts.igem.org/wiki/index.php/Part:BBa_T9002 BBa_T9002] | *An inducible gene expression device that is well-characterized in the registry [https://parts.igem.org/wiki/index.php/Part:BBa_T9002 BBa_T9002] | ||
<br> | <br> | ||
| Line 67: | Line 91: | ||
* To understand better some of the specificities of this new chassis we have built a basic model. | * To understand better some of the specificities of this new chassis we have built a basic model. | ||
* Our study investigates the mechanism of constitutive gene expression, inside a cell-free system with limited resources. | * Our study investigates the mechanism of constitutive gene expression, inside a cell-free system with limited resources. | ||
| − | * [[ | + | * [[Help:Cell-free chassis/Modelling| Read more ]] about our model. |
| + | <br> | ||
==Cell-Free Systems investigated== | ==Cell-Free Systems investigated== | ||
Different compartmentalization strategies have been explored to prolong the expression lifespan of the CFS. | Different compartmentalization strategies have been explored to prolong the expression lifespan of the CFS. | ||
| + | {| border="0" width="100%" | ||
| + | |- | ||
| + | ||[[Image:Batch_Mode.JPG]]|| | ||
*'''Batch mode CFS:''' Transcription-translation reaction is carried out in bulk solution. Expression is usually limited by nutrient (nucleotides and amino acids) and energy supplies. | *'''Batch mode CFS:''' Transcription-translation reaction is carried out in bulk solution. Expression is usually limited by nutrient (nucleotides and amino acids) and energy supplies. | ||
| + | |- | ||
| + | ||[[Image:Continuous_Exchange.JPG|100px]]|| | ||
*'''Continuous-exchange:''' Transcription-translation reaction is separated from the feeding solution by a dialysis membrane. Expression is sustained by diffusion of nutrients from the feeding soltuion to the reaction, and wastes generated by the reaction is diluted in the feeding solution. | *'''Continuous-exchange:''' Transcription-translation reaction is separated from the feeding solution by a dialysis membrane. Expression is sustained by diffusion of nutrients from the feeding soltuion to the reaction, and wastes generated by the reaction is diluted in the feeding solution. | ||
| + | |- | ||
| + | ||[[Image:Vesicle.JPG]]|| | ||
*'''Vesicle-encapsulated CFS:''' The reaction is separated from the feeding solution by a phospholipid bilayer. Expression is maintained for a longer time period than batch-mode CFS because of exchange of materials between the reaction and the feeding solution across the membrane. More reliable exchange of materials is established by inserting a non-specific pore protein with a suitable channel size into the phospholipid bilayer.<cite>3</cite> | *'''Vesicle-encapsulated CFS:''' The reaction is separated from the feeding solution by a phospholipid bilayer. Expression is maintained for a longer time period than batch-mode CFS because of exchange of materials between the reaction and the feeding solution across the membrane. More reliable exchange of materials is established by inserting a non-specific pore protein with a suitable channel size into the phospholipid bilayer.<cite>3</cite> | ||
| + | |- | ||
| + | |} | ||
Please click on each system for its individual characterization. | Please click on each system for its individual characterization. | ||
{| border="0" width="100%" | {| border="0" width="100%" | ||
|- align=center | |- align=center | ||
| − | |||
||[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30 <font face=georgia color=#330033 size=4>Commercial ''E. coli'' S30</font>] | ||[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30 <font face=georgia color=#330033 size=4>Commercial ''E. coli'' S30</font>] | ||
||[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_T7_S30 <font face=georgia color=#330033 size=4>Commercial ''E. coli'' T7 S30</font>] | ||[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_T7_S30 <font face=georgia color=#330033 size=4>Commercial ''E. coli'' T7 S30</font>] | ||
| + | ||[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Homemade_E.coli_S30 <font face=georgia color=#330033 size=4>Homemade ''E. coli'' S30</font>] | ||
||[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Vesicle <font face=georgia color=#330033 size=4>Vesicle-encapsulation</font>] | ||[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Vesicle <font face=georgia color=#330033 size=4>Vesicle-encapsulation</font>] | ||
|- | |- | ||
|} | |} | ||
| + | <br> | ||
==References== | ==References== | ||
Latest revision as of 22:58, 1 February 2019
Cell-Free Systems
Please click on each system for its individual characterization.
| Commercial E. coli S30 | Commercial E. coli T7 S30 | Homemade E. coli S30 | Vesicle-encapsulation |
Introduction
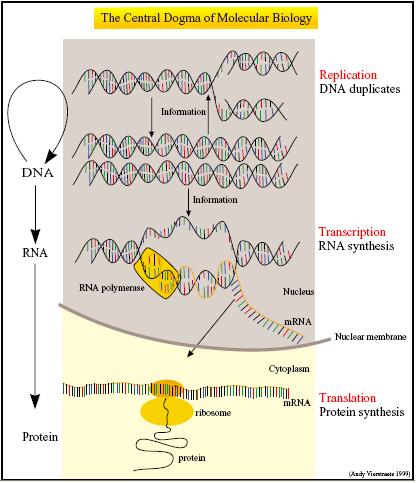
Cell-Free Systems (CFS) involve the in-vitro expression of genes into proteins. These systems can serve as a compatible chassis for the various parts and devices from the Registry of Standard Biological Parts.
The Central Dogma describes gene expression in terms of two essential processes - the transcription of DNA into messenger RNA (mRNA) and the translation of mRNA into polypeptides. 1 Not only do CFS house the molecular machinery necessary for transcription and translation, they are also optimized for these two processes.
Coupled transcription-translation systems usually combine a bacteriophage RNA polymerase and promoter with eukaryotic or prokaryotic extracts rich in ribosomes, transfer RNAs and aminoacyl-tRNA transferase enzymes. Buffers are also added to maintain the appropriate magnesium and salt concentrations required for efficient translation. In addition, an ATP regenerating system involving either creatine phosphate and creatine kinase or phosphoenolpyruvate and pyruvate kinase is used to power and prolong the lifespan of the expression machinery 2.
A good analogy compared this transcription-translation machinery of the CFS to the hardware and the synthetic DNA to the software. 3 This gives an apt illustration of the ease of building genetically engineered machines using the cell-free approach. Simply by adding the DNA template to the cell extract and feeding solution, the CFS would be able to express the encoded genetic circuit.
The PURE system has recently been developed as a reconstituted CFS for synthesizing proteins using recombinant elements 4. This purely synthetic expression system enables even better quality control over the reaction conditions.
Advantages and disadvantages of CFS
| System is not an organism and is not restricted by the policies imposed on genetically modified organisms(GMO) | Short expression lifespan because of limited energy of the system even in the presence of an ATP regenerating system |
| Process is quick and simple requiring only preparation of cell extract and feeding solution and subsequent addition of DNA template | Expensive system has no sustained metabolism to convert cheap energy (like sugars) into useable one for the gene expression machinery |
| No concurrent expression of existing genome, therefore your genetically engineered device is more energy efficient | Less characterization and experience of use in the laboratories compared to E. coli |
| No DNA mutation of your genetically engineered device because there is no DNA replication | |
| No selective pressue on your genetically engineered device because the system is non-living and does not undergo natural selection | |
| No self-replication of your genetically engineered device leads to a fixed amount of DNA being expressed and more control over the rate of expression | |
| Expression system can be quality-controlled by manipulating adjustable parameters e.g. buffers are added to maintain optimum magnesium concentrations for efficient translation; protease inhibitors can be added to minimize degradation of synthesized proteins |
Specifications for CFS characterization
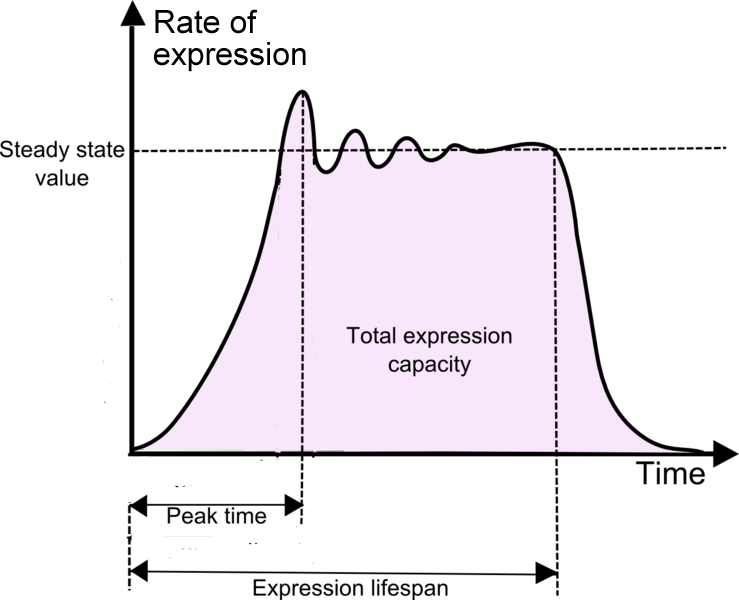
Several parameters are identified to help us understand the advantages and disadvantages associated with using a particular chassis.
| Properties | |
| Basic criterion for a chassis | Ability to express Biobricks |
| Temperature dependence | The relationship between expression rate and temperature. |
| Optimum amount of DNA | Amount of DNA required for expression rate to reach a maximum value. |
| Product stability | Measure of the half-life of a given protein (e.g. GFP) synthesized in the chassis. |
| Peak time Expression lifespan |
Measure of time from start of reaction to the point when expression rate reaches the maximum value Measure of time from start of reaction to the point when protein degradation overrides protein synthesis |
| Expression capacity | Measure of the total expression of a chassis for a given DNA construct template. This should take into account the degradation of synthesized protein. |
To gain knowledge about the CFS, we have selected the following parts available in the registry to extract the above-mentioned properties.
- A simple constitutive gene expression device that requires an E. coli RNA polymerase BBa_I13522
- A simple constitutive gene expression device that requires a T7 bacteriophage RNA polymerase BBa_I719015
- An inducible gene expression device that is well-characterized in the registry BBa_T9002
Cell-Free Systems modelling
- To understand better some of the specificities of this new chassis we have built a basic model.
- Our study investigates the mechanism of constitutive gene expression, inside a cell-free system with limited resources.
- Read more about our model.
Cell-Free Systems investigated
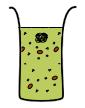
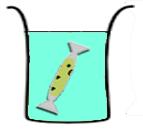
Different compartmentalization strategies have been explored to prolong the expression lifespan of the CFS.
Please click on each system for its individual characterization.
| Commercial E. coli S30 | Commercial E. coli T7 S30 | Homemade E. coli S30 | Vesicle-encapsulation |
References
<biblio>
- 1 pmid=4913914
- 2 pmid=15183761
- 3 pmid=16224117
- 4 pmid=16076456
- 5 pmid=14559971
</biblio>