Difference between revisions of "Part:BBa K2278021"
| (42 intermediate revisions by 2 users not shown) | |||
| Line 6: | Line 6: | ||
=='''Introduction'''== | =='''Introduction'''== | ||
| − | + | ||
This DNA biobrick was designed in order to produce an antimicrobial peptide from the crocodile <i>Crocodylus siamensis</i>. | This DNA biobrick was designed in order to produce an antimicrobial peptide from the crocodile <i>Crocodylus siamensis</i>. | ||
<h3 id="RT"> 1- Biological background </h3> | <h3 id="RT"> 1- Biological background </h3> | ||
| − | |||
Antimicrobial peptides (AMP) are phylogenetically ancient components of the innate defense of both invertebrates and vertebrates. In the context of growing bacterial antibiotic-resistance, these AMP are considered as potential new therapeutical candidates. | Antimicrobial peptides (AMP) are phylogenetically ancient components of the innate defense of both invertebrates and vertebrates. In the context of growing bacterial antibiotic-resistance, these AMP are considered as potential new therapeutical candidates. | ||
| − | |||
| − | |||
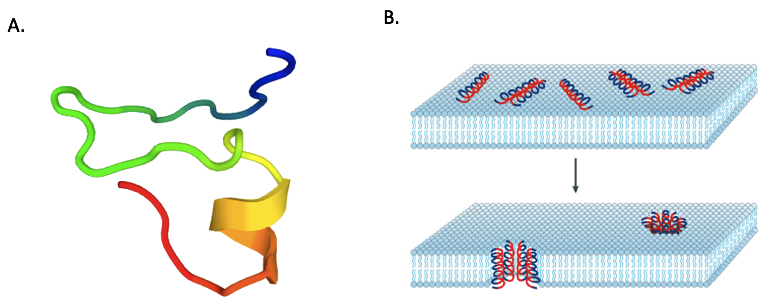
| + | The D-NY15 construction was based on the small peptide Leucrocin I sequence (NGVQPKY ; Yaraksa <i>et al</i>., 2013). Leucrocin I (<partinfo>BBa_K2278022</partinfo>) comes from Siamese crocodile white blood cells and shows a good antibacterial activity towards <i>Vibrio cholerae</i>. The sequence of Leucrocin I was improved to enhance the antimicrobial properties which depends on net charge (the AMP binds to a negatively charged membrane), length, structure, and hydrophobic percentage (to insert and permeabilize the microbial membrane). To do that Yaraksa <i>et al.</i> added hydrophobic amino acids (A4, L6, F7, V8, F11) and positively charged amino acids (K2, K3, K13) to Leucrocin I sequence. This results in D-NY15 (NKKAGLFVVQFPKKY ; figure 1). | ||
| + | |||
| + | |||
| + | [[Image:T--INSA-UPS_France--NY-15.png|800px|thumb|center|'''Figure 1:''' <b> (A) 3D predicted structure of NY-15 antimicrobial peptide </b> modeled on line with Mobyle portal. <b>(B) Example of an AMP model of intracellular killing mechanism</b> : the barrel-stave model of antimicrobial-peptide-induced killing (Brogden, 2005)]] | ||
| − | |||
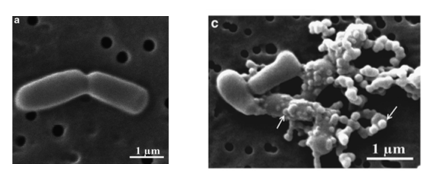
The mechanism of action of D-NY15 has been observed by transmission electron microscopy. The AMP binds and inserts to bacterium membranes to create pores, leading to the lysis of the cells (figure 2). | The mechanism of action of D-NY15 has been observed by transmission electron microscopy. The AMP binds and inserts to bacterium membranes to create pores, leading to the lysis of the cells (figure 2). | ||
| − | |||
| − | < | + | [[Image:T--INSA-UPS_France--Yaraksa-kun.png|800px|thumb|center|'''Figure 2:'''<b>Scanning electron micrographs of Vibrio cholerae treated with peptides </b> (a) control bacteria, (c) bacteria treated with D-NY15 AMP (Yaraksa <i>et al</i>., 2014)]] |
| − | |||
| + | <h3 id="RT"> 2- Usage in iGEM projects </h3> | ||
| − | </html> | + | The part was designed during the Croc’n Cholera project <html><a href="http://2017.igem.org/Team:INSA-UPS_France">(team INSA-UPS-France 2017)</a></html>. It contains the coding sequence to produces the D-NY15 AMP with the yeast pGAP promoter. The α-factor (<partinfo>BBa_K1800001</partinfo>) sequence contains a RBS and a signal sequence to secrete the produced peptides. |
=='''Experiments'''== | =='''Experiments'''== | ||
| − | + | ||
<h3 id="RT"> 1- Molecular biology </h3> | <h3 id="RT"> 1- Molecular biology </h3> | ||
| − | |||
| − | |||
| − | The construct was cloned by conventional ligation into pSB1C3 plasmid | + | The gene was placed under the control of an alpha factor signal. IDT performed the DNA synthesis and delivered the part as gBlock. The construct was cloned by conventional ligation into the pSB1C3 plasmid. |
| − | + | ||
| − | + | ||
| − | + | ||
<b>Analysis of the restriction map </b> | <b>Analysis of the restriction map </b> | ||
| − | + | [[Image:T--INSA-UPS_France--D-NY15.png|800px|thumb|center|'''Figure 3:''' <b>Analysis of the restriction map BBa_K2278021. </b> Digested fragments (Xba1 and Pst1) are electrophoresed through a 1% agarose gel. Control vector pSB1C3 contained an insert and expected size were 2034 and 700 bp (digestion was not total, hence the 2734 bp fragment). The fragment lengths of the 4 tested clones were 2035 bp and 343 bp for the required insert.]] | |
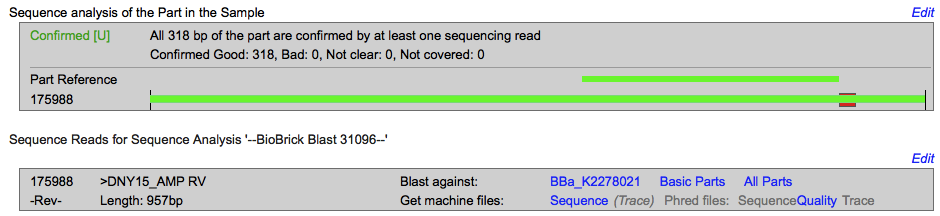
| − | + | <b>Sequencing </b> | |
| − | + | [[Image:T--INSA-UPS_France--NY15Seq.png|800px|thumb|center|'''Figure 4:''' <b>Sequencing of pSB1C3-D-NY15 </b> 1500 ng of plasmid are sequenced. The obtained sequence were blast on the BBa_K2278021 sequence with the iGEM sequencing online tools. ]] | |
The sequencing successfully validated the sequence of the biobrick. | The sequencing successfully validated the sequence of the biobrick. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | <h3 id="RT"> 2- Integration in <i>Pichia pastoris</i> </h3> | |
| + | The biobrick was placed under the control of the constitutive pGAP promoter (<partinfo>BBa_K431009</partinfo>) and was cloned in the pPICZalpha vector, an expression vector for the yeast <i>Pichia pastoris</i>. | ||
| − | < | + | The plasmid was then linearized and transferred in <i>Pichia pastoris</i> by electroporation. The integration is done by using pAOXI homology region (present on pPICZalpha). Indeed, the pAOXI promoter made genome recombination easier in <i>Pichia pastoris</i> than pGAP. |
| − | < | + | |
| − | |||
| − | |||
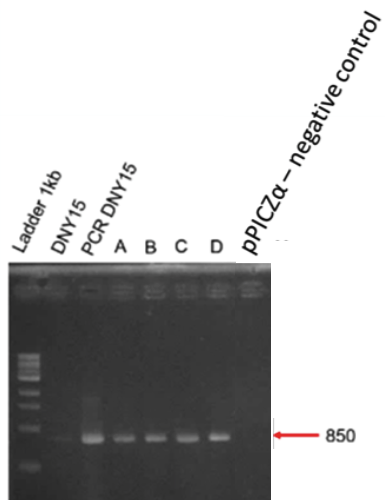
| + | [[Image:T--INSA-UPS_France--NY15inté.png|800px|thumb|center|'''Figure 5:''' <b>Integration of pGAP+BBa_K2278021 in <i>Pichia pastoris</i> </b> To verify the correct integration, we performed colony PCR and electrophoresed the product through a 1% agarose gel. A 850 bp fragment is expected if the integration is correct. ]] | ||
| − | + | Correct amplifications were observed for the 4 colonies tested (A to D) and the positive controls with the D-NY15 fragment as matrix (D-NY15) or the pPICZalpha-D-NY15 plasmid (PCR D-NY15). Negative control with pPICZalpha presented no band, as expected. | |
| + | |||
=='''Characterization'''== | =='''Characterization'''== | ||
| − | |||
| − | <h3 id="RT">1- Validation | + | D-NY15 production was performed with <i>Pichia pastoris</i> in YPD 40 g/L glucose grown for 4 days at 30 °C with shacking. Cells and supernatants were sampled. |
| + | |||
| + | <h3 id="RT">1- Validation of D-NY15 expression in <i>Pichia pastoris</i> by qRT-PCR </h3> | ||
| − | < | + | RNA from the <i>Pichia pastoris</i> cells were extracted and qRT-PCR was performed (figure 6). |
| + | [[Image:T--INSA-UPS_France--RTQPCRdekalitaäy.png|800px|thumb|center|'''Figure 6:''' <b>RTq-PCR of D-NY15. </b>RNA were issued from <i>Pichia pastoris</i> strains with or without the D-NY15 encoding gene. Total RNAs were extracted from transformants and reverse transcriptions were performed using Superscript II reverse transcriptase (Invitrogen). Resulting D-NY15 cDNA were then amplified by quantitative PCR. The curves correspond to <i>P. pastoris</i> with integrated pPICZalpha-DNY15 (A1, A2, A3), water control (B1 & B2), and <i>P. pastoris</i> with integrated pPICZalpha (C1, C2, C3).]] | ||
| − | |||
| − | The | + | The amount of fluorescence provided by the qRT-PCR with the D-NY15 primers rose after 8 cycles whereas the negative control (pPICZalpha only) started to be amplified at over 29 cycles (non specific amplification). This means that the D-NY15 encoding gene is expressed in <i>Pichia pastoris</i>. |
| − | < | + | |
| + | <h3 id="RT">2. Toxicity assay </h3> | ||
| + | The supernatants of <i>Pichia pastoris</i> strains with or without the D-NY15 encoding gene were used in a halo assay against <i>V. harveyi</i> as the target of AMPs. Briefly, 35mL of supernatants were freeze-dried and then resuspended in 3.5mL of water. A paper cut was soaked with one of these solutions and placed on a Petri plate inoculated with <i>V. harveyi</i> (figure 7). | ||
| + | |||
| + | [[Image:T--INSA-UPS_France--HalosPichia.png|800px|thumb|center|'''Figure 7:''' <b>AMP halo assay.</b> Positive control was performed with chloramphenicol (25 g/L), the negative control was performed with the empty plasmid integrated in <i>P. pastoris</i>, the assay was performed with the plasmid containing BBa_K2278021 integrated in <i>P. pastoris</i>.]] | ||
| + | <b>Conclusion : </b> | ||
| + | Haloes are formed around the positive control and the patch containing supernatants from a D-NY15 expressing strain. This nicely demonstrated the capacity of D-NY15 to inhibit the growth of <i>Vibrio harveyi</i>. This experiment has been performed twice. | ||
| + | Here, we show both the capacity of <i>Pichia pastoris</i> to produce antimicrobial peptides and the efficiency of the crocodile peptide D-NY15 to inhibit <i>Vibrio harveyi</i> growth. | ||
| − | |||
| − | + | <b>Perspectives: </b> | |
| − | + | ||
To go further, inhibition test should be now performed in liquid assay to be closer to the conditions in which we want to use these strains. | To go further, inhibition test should be now performed in liquid assay to be closer to the conditions in which we want to use these strains. | ||
| − | + | ||
===Design Notes=== | ===Design Notes=== | ||
| − | |||
| − | < | + | Part <partinfo>BBa_K1800001</partinfo> Alpha-Factor Secretion Signal was used. |
===Source=== | ===Source=== | ||
| − | The peptides DNA sequence has been obtained by reverse | + | The peptides DNA sequence has been obtained by reverse translation of the amino acid sequence of the D-NY15 proposed by Yaraksa <i>et al</i>., 2013, which was determined by mass spectrometry analysis. |
| − | + | ||
| + | <br> | ||
===References=== | ===References=== | ||
Yaraksa, N., Anunthawan, T., Theansungnoen, T., Daduang, S., Araki, T., Dhiravisit, A. and Thammasirirak, S. (2013). Design and synthesis of cationic antibacterial peptide based on Leucrocin I sequence, antibacterial peptide from crocodile (Crocodylus siamensis) white blood cell extracts. The Journal of Antibiotics, 67(3), pp.205-212. | Yaraksa, N., Anunthawan, T., Theansungnoen, T., Daduang, S., Araki, T., Dhiravisit, A. and Thammasirirak, S. (2013). Design and synthesis of cationic antibacterial peptide based on Leucrocin I sequence, antibacterial peptide from crocodile (Crocodylus siamensis) white blood cell extracts. The Journal of Antibiotics, 67(3), pp.205-212. | ||
Brogden KA (2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria. Nat Rev Microbiol. 3:238-50. | Brogden KA (2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria. Nat Rev Microbiol. 3:238-50. | ||
Latest revision as of 10:52, 30 October 2017
D-NY15 Antimicrobial peptide with Alpha-Factor Secretion Signal
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 244
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Introduction
This DNA biobrick was designed in order to produce an antimicrobial peptide from the crocodile Crocodylus siamensis.
1- Biological background
Antimicrobial peptides (AMP) are phylogenetically ancient components of the innate defense of both invertebrates and vertebrates. In the context of growing bacterial antibiotic-resistance, these AMP are considered as potential new therapeutical candidates.
The D-NY15 construction was based on the small peptide Leucrocin I sequence (NGVQPKY ; Yaraksa et al., 2013). Leucrocin I (BBa_K2278022) comes from Siamese crocodile white blood cells and shows a good antibacterial activity towards Vibrio cholerae. The sequence of Leucrocin I was improved to enhance the antimicrobial properties which depends on net charge (the AMP binds to a negatively charged membrane), length, structure, and hydrophobic percentage (to insert and permeabilize the microbial membrane). To do that Yaraksa et al. added hydrophobic amino acids (A4, L6, F7, V8, F11) and positively charged amino acids (K2, K3, K13) to Leucrocin I sequence. This results in D-NY15 (NKKAGLFVVQFPKKY ; figure 1).
The mechanism of action of D-NY15 has been observed by transmission electron microscopy. The AMP binds and inserts to bacterium membranes to create pores, leading to the lysis of the cells (figure 2).
2- Usage in iGEM projects
The part was designed during the Croc’n Cholera project (team INSA-UPS-France 2017). It contains the coding sequence to produces the D-NY15 AMP with the yeast pGAP promoter. The α-factor (BBa_K1800001) sequence contains a RBS and a signal sequence to secrete the produced peptides.
Experiments
1- Molecular biology
The gene was placed under the control of an alpha factor signal. IDT performed the DNA synthesis and delivered the part as gBlock. The construct was cloned by conventional ligation into the pSB1C3 plasmid.
Analysis of the restriction map
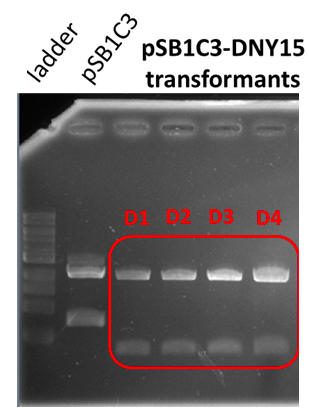
Sequencing
The sequencing successfully validated the sequence of the biobrick.
2- Integration in Pichia pastoris
The biobrick was placed under the control of the constitutive pGAP promoter (BBa_K431009) and was cloned in the pPICZalpha vector, an expression vector for the yeast Pichia pastoris.
The plasmid was then linearized and transferred in Pichia pastoris by electroporation. The integration is done by using pAOXI homology region (present on pPICZalpha). Indeed, the pAOXI promoter made genome recombination easier in Pichia pastoris than pGAP.
Correct amplifications were observed for the 4 colonies tested (A to D) and the positive controls with the D-NY15 fragment as matrix (D-NY15) or the pPICZalpha-D-NY15 plasmid (PCR D-NY15). Negative control with pPICZalpha presented no band, as expected.
Characterization
D-NY15 production was performed with Pichia pastoris in YPD 40 g/L glucose grown for 4 days at 30 °C with shacking. Cells and supernatants were sampled.
1- Validation of D-NY15 expression in Pichia pastoris by qRT-PCR
RNA from the Pichia pastoris cells were extracted and qRT-PCR was performed (figure 6).
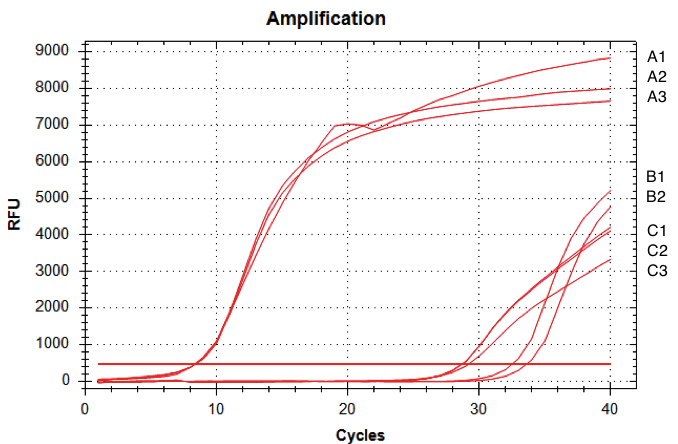
The amount of fluorescence provided by the qRT-PCR with the D-NY15 primers rose after 8 cycles whereas the negative control (pPICZalpha only) started to be amplified at over 29 cycles (non specific amplification). This means that the D-NY15 encoding gene is expressed in Pichia pastoris.
2. Toxicity assay
The supernatants of Pichia pastoris strains with or without the D-NY15 encoding gene were used in a halo assay against V. harveyi as the target of AMPs. Briefly, 35mL of supernatants were freeze-dried and then resuspended in 3.5mL of water. A paper cut was soaked with one of these solutions and placed on a Petri plate inoculated with V. harveyi (figure 7).
Conclusion :
Haloes are formed around the positive control and the patch containing supernatants from a D-NY15 expressing strain. This nicely demonstrated the capacity of D-NY15 to inhibit the growth of Vibrio harveyi. This experiment has been performed twice.
Here, we show both the capacity of Pichia pastoris to produce antimicrobial peptides and the efficiency of the crocodile peptide D-NY15 to inhibit Vibrio harveyi growth.
Perspectives:
To go further, inhibition test should be now performed in liquid assay to be closer to the conditions in which we want to use these strains.
Design Notes
Part BBa_K1800001 Alpha-Factor Secretion Signal was used.
Source
The peptides DNA sequence has been obtained by reverse translation of the amino acid sequence of the D-NY15 proposed by Yaraksa et al., 2013, which was determined by mass spectrometry analysis.
References
Yaraksa, N., Anunthawan, T., Theansungnoen, T., Daduang, S., Araki, T., Dhiravisit, A. and Thammasirirak, S. (2013). Design and synthesis of cationic antibacterial peptide based on Leucrocin I sequence, antibacterial peptide from crocodile (Crocodylus siamensis) white blood cell extracts. The Journal of Antibiotics, 67(3), pp.205-212.
Brogden KA (2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria. Nat Rev Microbiol. 3:238-50.