Part:BBa_K431009
glyceraldehyde 3-phosphate dehydrogenase promoter (pGAP)
This promoter is responsible for the transcription of glyceraldehyde 3-phosphate dehydrogenase in Pichia pastoris. Because this is a key enzyme for glycolysis, it is a strong constitutive promoter. This promoter is one of the most common alternatives to pAOX1 for heterologous protein production in P. pastoris. It is advantageous because its use obviates the hazards and expenses involved with using methanol and still provides high yields of protein.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 245
Team INSA-UPS France 2017: demonstration of pGAP validity in Pichia pastoris
We tested the functionality of pGAP as a constitutive promoter in the yeast Pichia pastoris background.
The biobrick was cloned in pPICZalpha vector and was integrated in Pichia pastoris at the pGAP genomic locus. The reporter gene here was BBa_K2278021 (D-NY15 antimicrobial peptide encoding gene for our project).
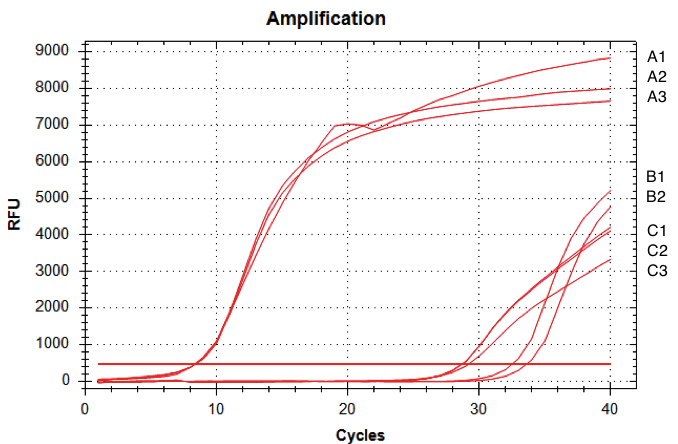
The amount of fluorescence provided by the qRT-PCR with the D-NY15 primers rose after 8 cycles whereas the negative control (pPICZalpha only) started to be amplified at over 29 cycles (non specific amplification). This means that the D-NY15 encoding gene is well expressed in Pichia pastoris with pGAP as a promoter. This promoter is therefore very efficient in the Pichia pastoris background.
Team SCU-China 2020: demonstration of pGAP validity in Saccharomyces cerevisiae and compare its strength with pTEF1
To demonstrate the validity of pGAP in Saccharomyces cerevisiae and choose the appropriate promoter for NLS-Csy4 and function genes, we measure the fluorescence intensity, and the data was divided by OD.
Figure 1: Relative fluorescence intensity of two fluorescence proteins under two promoters.
The signal of pGAP-CRISPR(Csy4)-DsRed-tADH1 is low, this is related to our plasmid design. We use double enzyme digestion and T4 ligation to construct pGAP-NLS-yeGFP-28nt-DsRed, the first step is the construction of pGAP-28nt-DsRed, which is called pGAP-R here. The secondary structure of 28nt may influence the expression level of DsRed.
In short, pGAP can work in Saccharomyces cerevisiae,and pTEF1 is stronger than pGAP (judged by the fluorescence of GFP and DsRed), so we use pTEF1 to express NLS-Csy4 for high cutting efficiency, and to design the construction of function genes additionally according to our modeling result.
//promoter
| None |

