Difference between revisions of "Part:BBa K864400"
(→Characterization of promoter in E.coli F' driving the extracellulare expression of dehalogenase DeHa2) |
|||
| (85 intermediate revisions by 12 users not shown) | |||
| Line 6: | Line 6: | ||
[1] Proc. Natl. Acad. Sci. USA, Vol. 80, pp. 21-25 | [1] Proc. Natl. Acad. Sci. USA, Vol. 80, pp. 21-25 | ||
| + | <html> | ||
| + | <h1>Contribution by TU_Kaiserslautern 2020</h1> | ||
| + | <p> | ||
| + | <b><u>Testing the influence of different IPTG-Concentrations on the induction of the tac-promotor in Escherichia coli</u></b><br> | ||
| + | The tac promotor we used is an established strong <i>E. coli</i> promotor. The promotor is a hybrid between the trp and lac UV5 promotors (BioCat GmbH). Replacing the -35 region of the lac UV5 promotor with the -35 region of the stronger trp promotor makes this hybrid way more efficient (de Boer et al., 1983). In addition to the -35 region the tac promotor also has a Pribnow box sequence to increase the efficiency (de Boer et al., 1983). The activity of the tac promotor can be repressed by a lac repressor and activated by inducing with isopropyl beta-D-thiogalactoside (IPTG), so a high regulation is possible (de Boer et al., 1983). The iGEM team iGEM12_Uppsala introduced the part BBa_K864400 to iGEM first in 2012. It was improved by Evry_Paris-Saclay in 2017 and characterized by by SHSBNU_China in 2019. Our goal was to verify this data.<br> | ||
| + | </p><p> | ||
| + | We wanted to help iGEM creating a big data bank of parts, because getting the greatest possible knowledge is everything in science. We worked with the tac promotor the whole time in E. coli to express the genetically code of our protein. So, we wanted to know, which influence the IPTG concentration has on the activity of the promotor. We transformed the pGEx-6P-1_baLac vector in E. coli AD494 (DE3) competent cells. For selection we used Kanamycin and Ampicillin resistance. </p><p> | ||
| + | We induced six liquid cultures of E.coli AD494 (DE3) pGEX-6P-1_baLac with the same optical density at 600 nm with different IPTG concentrations to compare different influences on the expression of the gene for the protein BaLac (Table 1). The cells grew at 17°C for 19 h. We took a sample before we induced and after expression for 19 h. We lysed the latter to separate the pellet and the soluble fraction with SDS-PAGE (Fig. 1).<bR></p> | ||
| + | <p> | ||
| + | <b>Table 1: Concentration of the IPTG in the six different pistons. </b> | ||
| + | </p> | ||
| + | <center><table > | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td>Piston </td> | ||
| + | <td>Concentration IPTG [mM] </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1 </td> | ||
| + | <td>0.250 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2 </td> | ||
| + | <td>0.375 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>3 </td> | ||
| + | <td>0.500 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>4 </td> | ||
| + | <td>1.000 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>5 </td> | ||
| + | <td>1.500 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>6 </td> | ||
| + | <td>2.500 </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table></center><br><br> | ||
| + | <p> | ||
| + | (A)<br> | ||
| + | <img src="https://2020.igem.org/wiki/images/a/a5/T--TU_Kaiserslautern--GEL_CO1.png"><br> | ||
| + | (B)<br> | ||
| + | <img src="https://2020.igem.org/wiki/images/1/10/T--TU_Kaiserslautern--GEL_CO2.png"><br> | ||
| + | <p> | ||
| + | <b>Fig. 1: SDS-PAGE of the test expression with different IPGT concentrations.</b> We induced E.coli AD494 (DE) pGEX-6P-1_baLac with six different IPTG concentrations. (A) shows the IPTG concentrations 1 3 (1: 0,25 mM; 2: 0,375 mM; 3: 0,5 mM), (B) shows the IPTG concentrations 4 6 (4: 1 mM; 5: 1,5 mM; 6: 2,5 mM). Samples were taken before induction and after induction expressing for 19 h. The latter were disrupted by sonication and insoluble and soluble fraction were separated. The red boxes show the produced translation fusion protein (BaLac and GST). The western blot was detected by anti-GST-antibodies (first antibody) and anti-Goat alkaline phosphatase conjugated antibodies (second antibody). Marker: New England BioLabs ® Blue Protein Standard Broad Range.<br></p> | ||
| + | <p> | ||
| + | In summary, we can say, that with increasing concentration of IPTG the production of protein increases as well (Fig. 1). That means, that there must have been a higher expression of the gene of interest (balac and gst). But you have to keep in mind, that sometimes E. coli has problems to fold foreign proteins correctly. This gets even worse if the expression of this protein is high. Figure 1 (B) shows, that the more IPTG is used, the more protein is in the insoluble fraction (pellet). That’s the reason why an IPTG concentration of 0.5 mM has established itself in our protein production – there we got the highest yield of soluble protein with the lowest concentration of IPTG.</p> | ||
| + | <u><b>References</b></u><bR><p> | ||
| + | de Boer HA, Comstock LJ, Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci U S A. 1983 Jan;80(1):21-5. doi: 10.1073/pnas.80.1.21. PMID: 6337371; PMCID: PMC393301.<br> | ||
| + | BioCat GmbH | ||
| + | </p> | ||
| + | |||
| + | </html> | ||
| + | |||
| + | <h1>Contribution by Uni Padua Team</h1> | ||
| + | ===Characterization of promoter in E.coli F' driving the extracellulare expression of dehalogenase DeHa2=== | ||
| + | <p>We used pTAC as a basic part in an expression cassette combining RBS (BBa_B003) and the coding sequence that encodes a carrier protein and the Dehalogenase type II gene (BBa_K5109016) | ||
| + | We tested the expression in TOP10 F’ Escherichia coli cells, since they keep the protein expression constantly repressed in basic conditions, due to the presence of lacI expression cassette in the F' episome. We then performed growth test of the transformed cells containing the expression cassette BBa_K5109023 compared to the growth of wild type TOP10 F’ cells. | ||
| + | Cellular growth was measured using a plate reader by taking the OD600 value every five minutes for 14 hours. | ||
| + | We then used the matrix of data generated by the plate reader to create the growth curves and analyze how they changed in the different concentrations of IPTG: respectively, 0.5 uM, 5uM, 50uM and 500 uM IPTG. | ||
| + | |||
| + | Overall, cellular growth was lower under IPTG induction compared to wild type colonies. | ||
| + | When analysing the data obtained from the growth rates for DeHa2, increasing the concentration of IPTG leads to a slow but constant decrease in the growth of the colonies. Further investigation is needed to decide the best IPTG concentration suitable in order to use our composite part. | ||
| + | |||
| + | <code><html><img src = "https://static.igem.wiki/teams/5109/c.png"></html></code> | ||
| + | |||
| + | <code><html><img src = "https://static.igem.wiki/teams/5109/b.png"></html></code> | ||
| + | |||
| + | <code><html><img src = "https://static.igem.wiki/teams/5109/a.png"></html></code> | ||
| + | |||
| + | ===Improvement by Evry_Paris-Saclay 2017=== | ||
| + | This part has been improved to be the pPsiTac1, a strong psicose inducible promoter. For more information on the improved part, please go to the page of [[Part:BBa_K2448016]]. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | |||
<!-- --> | <!-- --> | ||
| − | + | ===Characterization by SHSBNU_China 2019=== | |
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | |||
| + | <html> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <h3 id="CBD">Improvement</h3> | ||
| + | This part is improved by SHSBNU_China 2019 as <a href="https://parts.igem.org/Part:BBa_K3156000">BBa_K3156000</a> | ||
| + | <h3 id="CBD">Description</h3> | ||
| + | |||
| + | <body> | ||
| + | <p>In our design, the induction signal will be detected and stored in the plasmid DNA sequence of our genetically modified E. coli. The bacteria will be gathered from the capsule after it left human body and sent to lab for further quantitative analysis in order to represent the inflammation level in colon. Therefore, in our project, the fluorescence intensity will be the index that represents levels of gut inflammation. <br>Since then, we first needed to develop a standard quantitative measurement protocol, so we studied the following articles: Zong, Y et al (2017)[1] and Zhang, H. M. et al(2015). </p> | ||
| + | |||
| + | <figure> | ||
| + | <p style="text-align:center;"><img src="https://2019.igem.org/wiki/images/7/7f/T--SHSBNU_China---gfp_zqd.png" width="800" height="100"/> | ||
| + | <figcaption> | ||
| + | <b>Figure 1.We constructed our reporter circuit with sfgfp and pTac promoter</b> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | <p>Following reference, we determined the experimental procedure:<br> | ||
| + | Flat-bottom 96-well plates and sealing film were used throughout the study. Bacteria harboring parts/circuits of interest were inoculated from plates to LB medium and grown overnight (8−12 h, 1000 rpm, 37 °C, mB100-40 Thermo Shaker). Ten microliters of each overnight culture was sequentially diluted into 130 μL of fresh medium twice; the total dilution fold was 196. After growing the diluted cultures for ∼3 h, we diluted the exponentially growing cultures 700-fold using fresh medium; the dilution process was as follows: 10 μL of cell culture is added to 130 μL of M9 medium, which is followed by diluting 3 μL of this into 147 μL. Then, cultivation continued (1000 rpm, 37 °C, mB100-40); atspecifictimepoints,a2−50μL aliquot of each culture was transferred to a new plate containing 200 μL of PBS with 2 mg/mL kanamycin preadded to terminate protein expression. For the time course of cell growth after 700-fold dilution, OD 600 was recorded using Varioskan Flash (Thermal Scientific); the time interval was 5 min. | ||
| + | |||
| + | <figure> | ||
| + | <p style="text-align:center;"><img src="https://2019.igem.org/wiki/images/4/49/T--SHSBNU_China---characterization-step.jpeg" width="800" height="200"/> | ||
| + | <figcaption> | ||
| + | <b>Figure 2.Experimental procedure for recording the time course of gene expression using flow cytometry.</b> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | <p>The graph below shows our result.</p> | ||
| + | <figure> | ||
| + | <p style="text-align:center;"><img src="https://2019.igem.org/wiki/images/e/e5/T--SHSBNU_China---Ptac-result.png" width = "900" height ="400"/> | ||
| + | <figcaption> | ||
| + | |||
| + | <b>Figure 1.Ptac response curve in medium copy number vector.</b> | ||
| + | |||
| + | </figcaption> | ||
| + | </figure> | ||
| + | <b>Reference</b> | ||
| + | <p> [1]Zong, Y., Zhang, H. M., Lyu, C., Ji, X., Hou, J., Guo, X., ... & Lou, C. (2017). Insulated transcriptional elements enable precise design of genetic circuits. Nature communications, 8(1), 52.<br> | ||
| + | [2]Zhang, H. M., Chen, S., Shi, H., Ji, W., Zong, Y., Ouyang, Q., & Lou, C. (2015). Measurements of gene expression at steady state improve the predictability of part assembly. ACS synthetic biology, 5(3), 269-273.</br></p> | ||
| + | |||
| + | <!--下面是注释 | ||
| + | |||
| + | 分节 =='''2. [[Part:part编号|part名字]] characterization'''== | ||
| + | |||
| + | 大标题 <h3 id="CBD">________________</h3> | ||
| + | |||
| + | 中标题 <h5 id="CBD">________________</h5> | ||
| + | 段落 <p>_________</p> | ||
| + | |||
| + | 带圈标题 | ||
| + | <ul style="list-style-type:circle"> | ||
| + | <li><b>____________________</b></li> | ||
| + | </ul> | ||
| + | |||
| + | 图 | ||
| + | <figure> | ||
| + | <p style="text-align:center;"> | ||
| + | <img src="________" width = "800"/> | ||
| + | <figcaption> | ||
| + | _____________________________________ 斜体 <i>_________</i> | ||
| + | _____________________________________ 下标 <sub>_______</sub> | ||
| + | _____________________________________ 加粗 <b>_________</b> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | |||
| + | 上面是注释--> | ||
| + | |||
| + | </html> | ||
| + | <span class='h3bb'>Sequence and Features</span><br> | ||
| + | <partinfo>BBa_K864400 short</partinfo></br> | ||
| + | <partinfo>BBa_K864400 SequenceAndFeatures</partinfo> | ||
=='''1. [[Part:BBa_K1934040|pTAC_RFP]] characterization'''== | =='''1. [[Part:BBa_K1934040|pTAC_RFP]] characterization'''== | ||
<html> | <html> | ||
<h3 id="CBD">Description</h3> | <h3 id="CBD">Description</h3> | ||
| − | <p> | + | <p>pTAC promoter was taken from part <a href="https://parts.igem.org/Part:BBa_K864400">BBa_K864400</a>. This promoter is a hybrid of two operons: the trp and lac operons. This promoter is inducible by IPTG and commonly used in <i>Escherichia coli</i> for overproduction of proteins. <i>E. coli</i> NM522 strain that we used in our lab constitutively produces the LacIq protein, a strong pTAC promoter repressor. However, in absence of IPTG, we observed a strong leakage when plating our <a href="https://parts.igem.org/Part:BBa_K1934000">BBa_K1934000</a> transformants. Therefore, we decided to put a RFP reporter ORF under control of the pTAC promoter to characterize the promoter-driven transcriptional noise. |
</p> | </p> | ||
| Line 31: | Line 175: | ||
<h5 id="CBD">Experimental design</h5> | <h5 id="CBD">Experimental design</h5> | ||
| − | The RFP coding sequence <a href="https://parts.igem.org/Part:BBa_E1010"> | + | The RFP coding sequence (<a href="https://parts.igem.org/Part:BBa_E1010">BBa_E1010</a>) was placed <i>in silico</i> under the control of the pTAC promoter (<a href="https://parts.igem.org/Part:BBa_K864400">BBa_K864400</a>), a strong RBS (<a href="https://parts.igem.org/Part:BBa_B0030">BBa_B0030</a>) and a bidirectional terminator (<a href="https://parts.igem.org/Part:BBa_B0011">BBa_B0011</a>). IDT performed the DNA synthesis and delivered the part as gBlock. The construct was cloned by conventional ligation into pSB1C3 plasmid and then transformed into <i>E. coli</i> NM522 strain. |
| − | In order to study the efficiency of pTAC promoter for | + | In order to study the efficiency of the pTAC promoter for the overproduction of proteins, recombinant clones were grown overnight in LB at 37°C in duplicate in three different induction conditions (IPTG concentrations): 0 mmol.L<sup>-1</sup>, 1 mmol.L<sup>-1</sup> and 5 mmol.L<sup>-1</sup>. OD<sub>600</sub> of each culture was measured every hour over six hours. |
| − | <i> | + | <i>E. coli</i> NM522 strain was grown overnight in LB at 37°C in the same three induction conditions as control. |
| Line 44: | Line 188: | ||
</ul> | </ul> | ||
| − | <p>We | + | <p>We studied the noise of the promoter by comparing the normalized fluorescence between the construction and the NM522 strain without any induction of IPTG.</p> |
| − | <figure><p style="text-align:center;"><img src="https://static.igem.org/mediawiki/2016/f/fc/INSA-Lyon_RFP-NM522.jpg" width = "800"/><figcaption><b>Figure 1. Fluorescence/ | + | <figure><p style="text-align:center;"><img src="https://static.igem.org/mediawiki/2016/f/fc/INSA-Lyon_RFP-NM522.jpg" width = "800"/><figcaption><b>Figure 1. Fluorescence/OD<sub>600</sub> comparison between NM522 with or without the plasmid (RFP). </b> Fluorescence of each sample was measured every hour over six hours with ChemiDoc Imaging system and was normalized by dividing by the OD<sub>600</sub>. After one hour, a significant expression difference was noticed; NM522 strain with plasmid revealed a higher protein expression. </figcaption></figure> |
| − | <p>An ANOVA was made to see if there was a time effect between the two populations. We | + | <p>An ANOVA was made to see if there was a time effect between the two populations. We obtained a p-value of 0.61, suggesting that time had no effect on pTAC-RFP expression in absence of IPTG (α<0.05). Given this result, we gathered data to analyze if there was a significant difference between the two strains. |
| − | A Student test was performed with the variance not equal. The p-value of 5.44*10<sup>-4</sup> | + | A Student test was performed with the variance not equal. The p-value of 5.44*10<sup>-4</sup> indicated that the strain carrying pTAC-RFP transcriptional fusion displayed a higher fluorescence than the control strain.</p> |
<ul style="list-style-type:circle"> | <ul style="list-style-type:circle"> | ||
| Line 55: | Line 199: | ||
</ul> | </ul> | ||
| − | <p> | + | <p>Then we studied the induction of the promoter by comparing the normalized fluorescence of the construction under the induction of [IPTG] = 0 and 1 mmol.L<sup>-1</sup>.</p> |
| − | <figure><p style="text-align:center;"><img src="https://static.igem.org/mediawiki/2016/b/bd/INSA-Lyon_0vs1.jpg" width = "800"/><figcaption><b>Figure 2. Fluorescence/ | + | <figure><p style="text-align:center;"><img src="https://static.igem.org/mediawiki/2016/b/bd/INSA-Lyon_0vs1.jpg" width = "800"/><figcaption><b>Figure 2. Fluorescence/OD<sub>600</sub> comparison between NM522 strains with plasmid (RFP) and with IPTG induction of 0 or 1 mmol.L<sup>-1</sup>. </b> Fluorescence of each sample was measured every hour over six hours with ChemiDoc Imaging system and was normalized by dividing by the OD<sub>600</sub>. After one hour, a significant expression difference was noticed; IPTG induction enables a higher protein expression rate. </figcaption></figure> |
| − | <p>An ANOVA test was made to see if there was a time effect between the two populations. | + | <p>An ANOVA test was made to see if there was a time effect between the two populations. A p-value of 0.21 indicated that time had no effect on the fluorescence induction (α<0.05). Data was therefore gathered in order to compare the strain fluorescence with 1 mmol.L<sup>-1</sup> IPTG and no IPTG. |
| − | We | + | We realized a Student test with the variance not equal and obtained a p-value of 8.57*10<sup>-3</sup>, showing a difference of fluorescence due to the presence of IPTG in the medium.</p> |
<p>Finally, we compared the expression of the pTAC-RFP transcriptional fusion in 2 concentrations of IPTG: 1 and 5 mmol.L<sup>-1</sup>.</p> | <p>Finally, we compared the expression of the pTAC-RFP transcriptional fusion in 2 concentrations of IPTG: 1 and 5 mmol.L<sup>-1</sup>.</p> | ||
| − | <figure><p style="text-align:center;"><img src="https://static.igem.org/mediawiki/2016/6/64/INSA-Lyon_1vs5.jpg" width = "800"/><figcaption><b>Figure 3. Fluorescence/ | + | <figure><p style="text-align:center;"><img src="https://static.igem.org/mediawiki/2016/6/64/INSA-Lyon_1vs5.jpg" width = "800"/><figcaption><b>Figure 3. Fluorescence/OD<sub>600</sub> comparison between NM522 strains with plasmid (RFP) and with IPTG induction of 1 or 5 mmol.L<sup>-1</sup> </b> Fluorescence of each sample was measured every hour over six hours with ChemiDoc Imaging system and was normalized by dividing by the OD<sub>600</sub>. Even after six hours of incubation, no significant expression difference was observed, protein expression rate was not correlated with the concentration of IPTG. </figcaption></figure> |
| − | <p>An ANOVA was made to see if there was a time effect between the two populations. | + | <p>An ANOVA was made to see if there was a time effect between the two populations. A p-value of 0.06 indicated that time had no effect (α<0.05). From there, we could gather the data to analyze if there was a significant difference between the two concentrations of IPTG.</p> |
| − | <p>We | + | <p>We realized a Student test with the variance not equal and obtained a p-value of 0.61, indicating that no significant difference of fluorescence was observed with the rise of IPTG concentration.</p> |
<h3 id="CBD">Conclusion</h3> | <h3 id="CBD">Conclusion</h3> | ||
| − | <p>In the case of RFP, the pTAC promoter | + | <p>In the case of RFP, the pTAC promoter seemed to not enable a gene tune because statistics showed that there was a significant noise, even in the absence of IPTG.</p> |
| − | + | </html> | |
| − | + | ||
| − | + | ||
| + | =='''2. [[Part:BBa_K1934050|pTAC_CFP]] characterization'''== | ||
<html> | <html> | ||
<h3 id="CBD">Description</h3> | <h3 id="CBD">Description</h3> | ||
| − | <p> | + | <p>pTAC promoter was taken from part <a href="https://parts.igem.org/Part:BBa_K864400">part BBa_K864400</a> . This promoter is a hybrid of two operons: the trp and lac operons.This promoter is inducible by IPTG and commonly used in <i>Escherichia coli</i> for overproduction of proteins. <i>Escherichia coli</i> NM522 strain that we used in our lab constitutively express the LacIq protein, a strong pTAC promoter repressor. However, in absence of IPTG, we observed a strong leakage when plating our <a href="https://parts.igem.org/Part:BBa_K1934000">BBa_K1934000</a> transformants. Therefore, we decided to put a CFP reporter ORF under control of the pTAC promoter to characterize the promoter-driven transcriptional noise. |
<h3 id="CBD">Expression of the pTAC-CFP fusion in presence of increasing amount of the inductor IPTG</h3> | <h3 id="CBD">Expression of the pTAC-CFP fusion in presence of increasing amount of the inductor IPTG</h3> | ||
| Line 83: | Line 226: | ||
<h5 id="CBD">Experimental design</h5> | <h5 id="CBD">Experimental design</h5> | ||
| − | The CFP coding sequence <a href="https://parts.igem.org/Part:BBa_E2020"> | + | The CFP coding sequence (<a href="https://parts.igem.org/Part:BBa_E2020">BBa_E2020</a>) was placed <i>in silico</i> under the control of the pTAC promoter (<a href="https://parts.igem.org/Part:BBa_K864400">BBa_K864400</a>), a strong RBS (<a href="https://parts.igem.org/Part:BBa_B0030">BBa_B0030</a>) and a bidirectional terminator (<a href="https://parts.igem.org/Part:BBa_B0011">BBa_B0011</a>). IDT performed the DNA synthesis and delivered the part as gBlock. The construct was cloned by conventional ligation into pSB1C3 plasmid and then transformed into <i>E. coli</i> NM522 strain. |
| − | In order to study the efficiency of pTAC promoter for | + | In order to study the efficiency of the pTAC promoter for the overproduction of proteins, recombinant clones were grown overnight in LB at 37°C in duplicate in three different induction conditions (IPTG concentrations): 0 mmol.L<sup>-1</sup>, 1 mmol.L<sup>-1</sup> and 5 mmol.L<sup>-1</sup>. OD<sub>600</sub> of each culture was measured every hour over six hours. |
| − | <i> | + | <i>E. coli</i> NM522 strain was grown overnight in LB at 37°C in the same three induction conditions as control. |
| Line 96: | Line 239: | ||
</ul> | </ul> | ||
| − | <p>We | + | <p>We studied the noise of the promoter by comparing the normalized fluorescence between the construction and the NM522 strain without any induction of IPTG.</p> |
| − | <figure><p style="text-align:center;"><img src="https://static.igem.org/mediawiki/parts/5/59/T--INSA-Lyon--CFPvsNM522.jpeg" width = "800"/><figcaption><b>Figure | + | <figure><p style="text-align:center;"><img src="https://static.igem.org/mediawiki/parts/5/59/T--INSA-Lyon--CFPvsNM522.jpeg" width = "800"/><figcaption><b>Figure 1. Fluorescence/OD<sub>600</sub> comparison between NM522 with or without the plasmid (CFP). </b> Fluorescence of each sample was measured every hour over six hours with ChemiDoc Imaging system and was normalized by dividing by the OD<sub>600</sub>. After one hour, a significant expression difference was noticed; NM522 strain with plasmid revealed a higher protein expression. </figcaption></figure> |
| − | <p>An ANOVA was made to see if there was a time effect between the two populations. We obtain a p-value of 0.96, suggesting that | + | |
| − | A Student test was performed with the variance not equal. The p-value of 0.10 | + | <p>An ANOVA was made to see if there was a time effect between the two populations. We obtain a p-value of 0.96, suggesting that time had no effect on pTAC-CFP expression in absence of IPTG (α<0.05). Given this result, we gathered data to analyze if there was a significant difference between the two strains. |
| + | A Student test was performed with the variance not equal. The p-value of 0.10 indicated that the strain carrying pTAC-CFP transcriptional fusion didn’t display a significant fluorescence difference with the control strain.</p> | ||
<ul style="list-style-type:circle"> | <ul style="list-style-type:circle"> | ||
| Line 107: | Line 251: | ||
</ul> | </ul> | ||
| − | <p> | + | <p>Then we studied the induction of the promoter by comparing the normalized fluorescence of the construction under the induction of [IPTG] = 0 and 1 mmol.L<sup>-1</sup>.</p> |
| − | <figure><p style="text-align:center;"><img src="https://static.igem.org/mediawiki/parts/0/02/T--INSA-Lyon--CFP_0vs1.jpeg" width = "800"/><figcaption><b>Figure | + | <figure><p style="text-align:center;"><img src="https://static.igem.org/mediawiki/parts/0/02/T--INSA-Lyon--CFP_0vs1.jpeg" width = "800"/><figcaption><b>Figure 2. Fluorescence/OD<sub>600</sub> comparison between NM522 strains with plasmid (CFP) and with IPTG induction of 0 or 1 mmol.L<sup>-1</sup>. </b> Fluorescence of each sample was measured every hour over six hours with ChemiDoc Imaging system and was normalized by dividing by the OD<sub>600</sub>. After one hour, a significant expression difference was noticed; IPTG induction enabled a higher protein expression rate. </figcaption></figure> |
| − | <p>An ANOVA test was made to see if there was a time effect between the two populations. | + | <p>An ANOVA test was made to see if there was a time effect between the two populations. A p-value of 0.05 indicated that time had no effect on the fluorescence induction (α<0.05). Data was therefore gathered in order to compare the strain fluorescence with 1 mmol.L<sup>-1</sup> IPTG and no IPTG. |
| − | We | + | We realized a Student test with the variance not equal and obtained a p-value of 4.64*10<sup>-10</sup>, showing a significant difference of fluorescence due to the presence of IPTG in the medium.</p> |
<p>Finally, we compared the expression of the pTAC-CFP transcriptional fusion in 2 concentrations of IPTG: 1 and 5 mmol.L<sup>-1</sup>.</p> | <p>Finally, we compared the expression of the pTAC-CFP transcriptional fusion in 2 concentrations of IPTG: 1 and 5 mmol.L<sup>-1</sup>.</p> | ||
| − | <figure><p style="text-align:center;"><img src="https://static.igem.org/mediawiki/parts/a/a4/T--INSA-Lyon--CFP_1vs5.jpeg" width = "800"/><figcaption><b>Figure | + | <figure><p style="text-align:center;"><img src="https://static.igem.org/mediawiki/parts/a/a4/T--INSA-Lyon--CFP_1vs5.jpeg" width = "800"/><figcaption><b>Figure 3. Fluorescence/OD<sub>600</sub> comparison between NM522 strains with plasmid (CFP) and with IPTG induction of 1 or 5 mmol.L<sup>-1</sup> </b> Fluorescence of each sample was measured every hour during six hours with ChemiDoc Imaging system and was normalized by dividing by the OD<sub>600</sub>. Even after six hours of incubation, no significant expression difference could be detected, protein expression rate was not correlated with the concentration of IPTG. </figcaption></figure> |
| − | <p>An ANOVA was made to see if there was a time effect between the two populations. | + | <p>An ANOVA was made to see if there was a time effect between the two populations. A p-value of 0.55 indicated that time had no effect (α<0.05). From there, we gathered data to analyze if there was a significant difference between the two concentrations of IPTG. |
| − | We | + | We realized a Student test with the variance not equal and obtained a p-value of 0.54, indicating that no significant difference of fluorescence was observed with the rise of IPTG concentration. |
</p> | </p> | ||
<h3 id="CBD">Conclusion</h3> | <h3 id="CBD">Conclusion</h3> | ||
| − | <p>In the case of CFP, the pTAC promoter | + | <p>In the case of CFP, the pTAC promoter seemed to enable a gene tune because there wasn’t a differential gene expression in absence of IPTG, so a significant noise wasn’t measured.</p> |
| + | </html> | ||
| − | + | ==Team JUN_China 2019: Fluorescent characterization of the Tac promoter== | |
| − | + | Tac promoter is a hybrid of two operons: the trp and lac operons and it is inducible by IPTG.In our project, we built a number of parts which also contains a large number of construct intermediates using the tac promoter. The tac promoter is critical in our entire project, so we used green and red fluorescent protein to characterize its effectiveness. At the same time, we explored the changes in expression intensity of green and red fluorescent protein under the control of tac promoter by IPTG inducing at different times. | |
| + | Experiment design: | ||
| − | < | + | We ligated the <i>eGFP</i> and <i>RFP</i> gene into the loop vector pZM1 (Ptac) containing the inducible tac promoter respectively and transfered <i>eGFP</i> and <i>RFP</i> into our chassis microorganism: <i>Corynebacterium glutamicum</i> F343 to carry out fermentation experiment. We added 1mmol/L IPTG to the fermentation broth at 0h, 1h, and 2h respectively and measured the fluorescence intensity and cell growth at different times to investigate whether the inducing capability of tac promoter was different at different induction times. |
| − | + | During the detection process, the cells were washed using PBS and the cells were diluted by appropriate multiples according to the measurement range of the instrument. The growth state and fluorescence intensity of the cells were detected using 96-well plates. | |
| − | + | The green fluorescent protein has an excitation wavelength of 488 nm, an emission wavelength of 517 nm. The red fluorescent protein excitation wavelength is 560 nm, and the emission wavelength is 630 nm. | |
| − | + | ||
| − | + | Result: | |
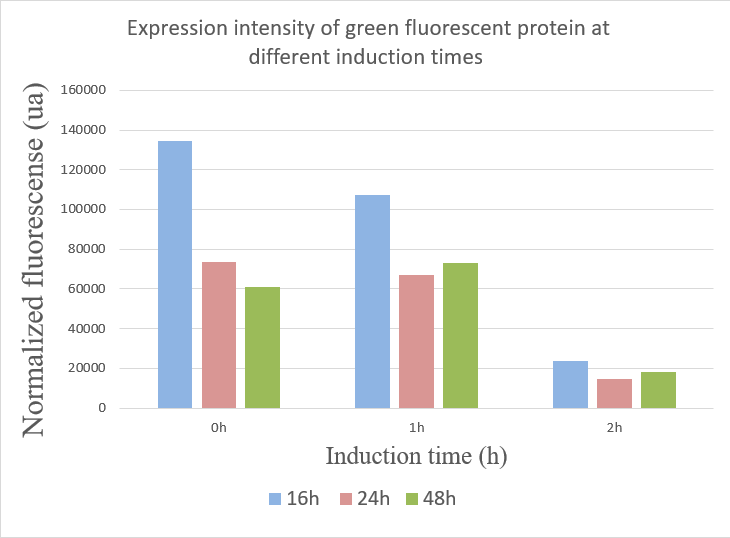
| − | + | We used the infinite 200Pro instrument to measure green and red fluorescent protein and OD600 measurement. The relevant data are shown below: | |
| − | + | [[image:Induce.png|400px]] | |
| − | + | ||
| − | + | The data showed: | |
| − | + | ||
| − | + | ||
| − | + | 1.When inducing at 0 hour, the highest fluorescence value were at 16 hours, and there was a gradual decrease in data from 24 hours to 48 hours. This may due to the degradation of fluorescent protein. | |
| − | + | 2.The fluorescence values induced at 1 hour were relatively stable, but when at 16 hours the fluorescence value was best. | |
| − | + | 3.The expression of GFP was the worst when inducing at 2 hour. | |
| − | + | ||
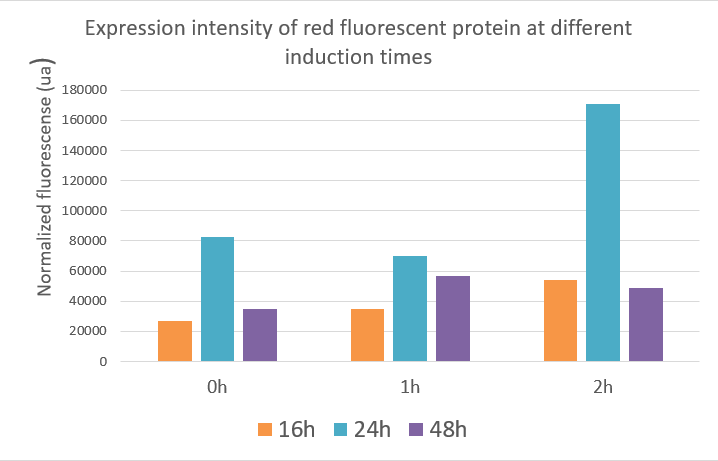
| − | + | [[image:Induce-2.png|400px]] | |
| − | + | ||
| + | Red fluorescent protein data showed that the expression of the red fluorescent protein was best when inducing at two hours and the value reached about 17,000 ua at 24 hours. The optimal induction time of red fluorescent protein expression was the same as the optimal induction time of polymerase in our project, which was induced at two hours after fermentation, and the product expression was highest. | ||
| + | The data indicated that induction at different times, the expression of the target gene and the yield of the product were different. | ||
| + | |||
| + | ==Team GreatBay_SZ 2020== | ||
| + | <html> | ||
| + | <p>We have used pTac to express the key protein of our project: <i>Geobacter sulfurreducens</i>' pilin-based electrically conductive protein nanowires (e-PNs). The gene circuit includes this tac promoter which promotes the pilin gene(<i>pilA</i>) to express protein monomers; the monomers are later assembled into protein nanowires by the TypeIV pilin assembly system, which is constructed from the gene cluster at the downstream of the pilin gene(figure 1). </p> | ||
| + | <div style="text-align: center;"> | ||
| + | <img src="https://2020.igem.org/wiki/images/6/64/T--GreatBay_SZ--circuit2.png" alt="" width="700"> | ||
| + | <h6 style="text-align:center">Figure 1: <strong>A</strong> Construction of BIOT gene circuit</h6> | ||
| + | </div> | ||
| + | <p>The e-PN can be successfully expressed by express this part under the control of this pTac, and the result was verified by Western Blot(figure 2). | ||
| + | </p> | ||
| + | <div style="text-align: center;"> | ||
| + | <img src="https://2020.igem.org/wiki/images/4/4e/T--GreatBay_SZ--western.png" alt="" width="400"> | ||
| + | <h6 style="text-align:center">Figure 2: Western Blot of the cell lysis of BIOT and BIOT-His(express e-PN and e-PN-His respectively); Antibody: Anti-His tag; e-PN-His: 7.7 kDa.</h6> </div> | ||
</html> | </html> | ||
| − | <!-- | + | |
| + | <!-- Add more about the biology of is part here.> | ||
| + | |||
| + | |||
| + | ==='''2019 SHSBNU China'''=== | ||
| + | |||
===Functional Parameters=== | ===Functional Parameters=== | ||
| − | <partinfo> | + | <partinfo>BBa_K2963032 parameters</partinfo> |
<!-- --> | <!-- --> | ||
Latest revision as of 13:39, 1 October 2024
Ptac, trp & lac regulated promoter
The Ptac promoter is a functional hybrid promoter, derived from the trp and lac promoters, that are regulated by trp and lac [1]. This part also exist together with lacI, part BBa_K180000
[1] Proc. Natl. Acad. Sci. USA, Vol. 80, pp. 21-25
Contribution by TU_Kaiserslautern 2020
Testing the influence of different IPTG-Concentrations on the induction of the tac-promotor in Escherichia coli
The tac promotor we used is an established strong E. coli promotor. The promotor is a hybrid between the trp and lac UV5 promotors (BioCat GmbH). Replacing the -35 region of the lac UV5 promotor with the -35 region of the stronger trp promotor makes this hybrid way more efficient (de Boer et al., 1983). In addition to the -35 region the tac promotor also has a Pribnow box sequence to increase the efficiency (de Boer et al., 1983). The activity of the tac promotor can be repressed by a lac repressor and activated by inducing with isopropyl beta-D-thiogalactoside (IPTG), so a high regulation is possible (de Boer et al., 1983). The iGEM team iGEM12_Uppsala introduced the part BBa_K864400 to iGEM first in 2012. It was improved by Evry_Paris-Saclay in 2017 and characterized by by SHSBNU_China in 2019. Our goal was to verify this data.
We wanted to help iGEM creating a big data bank of parts, because getting the greatest possible knowledge is everything in science. We worked with the tac promotor the whole time in E. coli to express the genetically code of our protein. So, we wanted to know, which influence the IPTG concentration has on the activity of the promotor. We transformed the pGEx-6P-1_baLac vector in E. coli AD494 (DE3) competent cells. For selection we used Kanamycin and Ampicillin resistance.
We induced six liquid cultures of E.coli AD494 (DE3) pGEX-6P-1_baLac with the same optical density at 600 nm with different IPTG concentrations to compare different influences on the expression of the gene for the protein BaLac (Table 1). The cells grew at 17°C for 19 h. We took a sample before we induced and after expression for 19 h. We lysed the latter to separate the pellet and the soluble fraction with SDS-PAGE (Fig. 1).
Table 1: Concentration of the IPTG in the six different pistons.
| Piston | Concentration IPTG [mM] |
| 1 | 0.250 |
| 2 | 0.375 |
| 3 | 0.500 |
| 4 | 1.000 |
| 5 | 1.500 |
| 6 | 2.500 |
(A)

(B)

Fig. 1: SDS-PAGE of the test expression with different IPGT concentrations. We induced E.coli AD494 (DE) pGEX-6P-1_baLac with six different IPTG concentrations. (A) shows the IPTG concentrations 1 3 (1: 0,25 mM; 2: 0,375 mM; 3: 0,5 mM), (B) shows the IPTG concentrations 4 6 (4: 1 mM; 5: 1,5 mM; 6: 2,5 mM). Samples were taken before induction and after induction expressing for 19 h. The latter were disrupted by sonication and insoluble and soluble fraction were separated. The red boxes show the produced translation fusion protein (BaLac and GST). The western blot was detected by anti-GST-antibodies (first antibody) and anti-Goat alkaline phosphatase conjugated antibodies (second antibody). Marker: New England BioLabs ® Blue Protein Standard Broad Range.
In summary, we can say, that with increasing concentration of IPTG the production of protein increases as well (Fig. 1). That means, that there must have been a higher expression of the gene of interest (balac and gst). But you have to keep in mind, that sometimes E. coli has problems to fold foreign proteins correctly. This gets even worse if the expression of this protein is high. Figure 1 (B) shows, that the more IPTG is used, the more protein is in the insoluble fraction (pellet). That’s the reason why an IPTG concentration of 0.5 mM has established itself in our protein production – there we got the highest yield of soluble protein with the lowest concentration of IPTG.
References
de Boer HA, Comstock LJ, Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci U S A. 1983 Jan;80(1):21-5. doi: 10.1073/pnas.80.1.21. PMID: 6337371; PMCID: PMC393301.
BioCat GmbH
Contribution by Uni Padua Team
Characterization of promoter in E.coli F' driving the extracellulare expression of dehalogenase DeHa2
We used pTAC as a basic part in an expression cassette combining RBS (BBa_B003) and the coding sequence that encodes a carrier protein and the Dehalogenase type II gene (BBa_K5109016)
We tested the expression in TOP10 F’ Escherichia coli cells, since they keep the protein expression constantly repressed in basic conditions, due to the presence of lacI expression cassette in the F' episome. We then performed growth test of the transformed cells containing the expression cassette BBa_K5109023 compared to the growth of wild type TOP10 F’ cells.
Cellular growth was measured using a plate reader by taking the OD600 value every five minutes for 14 hours.
We then used the matrix of data generated by the plate reader to create the growth curves and analyze how they changed in the different concentrations of IPTG: respectively, 0.5 uM, 5uM, 50uM and 500 uM IPTG.
Overall, cellular growth was lower under IPTG induction compared to wild type colonies.
When analysing the data obtained from the growth rates for DeHa2, increasing the concentration of IPTG leads to a slow but constant decrease in the growth of the colonies. Further investigation is needed to decide the best IPTG concentration suitable in order to use our composite part.



Improvement by Evry_Paris-Saclay 2017
This part has been improved to be the pPsiTac1, a strong psicose inducible promoter. For more information on the improved part, please go to the page of Part:BBa_K2448016.
Characterization by SHSBNU_China 2019
Improvement
This part is improved by SHSBNU_China 2019 as BBa_K3156000Description
In our design, the induction signal will be detected and stored in the plasmid DNA sequence of our genetically modified E. coli. The bacteria will be gathered from the capsule after it left human body and sent to lab for further quantitative analysis in order to represent the inflammation level in colon. Therefore, in our project, the fluorescence intensity will be the index that represents levels of gut inflammation.
Since then, we first needed to develop a standard quantitative measurement protocol, so we studied the following articles: Zong, Y et al (2017)[1] and Zhang, H. M. et al(2015).

Following reference, we determined the experimental procedure:
Flat-bottom 96-well plates and sealing film were used throughout the study. Bacteria harboring parts/circuits of interest were inoculated from plates to LB medium and grown overnight (8−12 h, 1000 rpm, 37 °C, mB100-40 Thermo Shaker). Ten microliters of each overnight culture was sequentially diluted into 130 μL of fresh medium twice; the total dilution fold was 196. After growing the diluted cultures for ∼3 h, we diluted the exponentially growing cultures 700-fold using fresh medium; the dilution process was as follows: 10 μL of cell culture is added to 130 μL of M9 medium, which is followed by diluting 3 μL of this into 147 μL. Then, cultivation continued (1000 rpm, 37 °C, mB100-40); atspecifictimepoints,a2−50μL aliquot of each culture was transferred to a new plate containing 200 μL of PBS with 2 mg/mL kanamycin preadded to terminate protein expression. For the time course of cell growth after 700-fold dilution, OD 600 was recorded using Varioskan Flash (Thermal Scientific); the time interval was 5 min.

The graph below shows our result.

[1]Zong, Y., Zhang, H. M., Lyu, C., Ji, X., Hou, J., Guo, X., ... & Lou, C. (2017). Insulated transcriptional elements enable precise design of genetic circuits. Nature communications, 8(1), 52.
[2]Zhang, H. M., Chen, S., Shi, H., Ji, W., Zong, Y., Ouyang, Q., & Lou, C. (2015). Measurements of gene expression at steady state improve the predictability of part assembly. ACS synthetic biology, 5(3), 269-273.
Ptac, trp & lac regulated promoter</br>
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
1. pTAC_RFP characterization
Description
pTAC promoter was taken from part BBa_K864400. This promoter is a hybrid of two operons: the trp and lac operons. This promoter is inducible by IPTG and commonly used in Escherichia coli for overproduction of proteins. E. coli NM522 strain that we used in our lab constitutively produces the LacIq protein, a strong pTAC promoter repressor. However, in absence of IPTG, we observed a strong leakage when plating our BBa_K1934000 transformants. Therefore, we decided to put a RFP reporter ORF under control of the pTAC promoter to characterize the promoter-driven transcriptional noise.
Expression of the pTAC-RFP fusion in presence of increasing amount of the inductor IPTG
Experimental design
The RFP coding sequence (BBa_E1010) was placed in silico under the control of the pTAC promoter (BBa_K864400), a strong RBS (BBa_B0030) and a bidirectional terminator (BBa_B0011). IDT performed the DNA synthesis and delivered the part as gBlock. The construct was cloned by conventional ligation into pSB1C3 plasmid and then transformed into E. coli NM522 strain. In order to study the efficiency of the pTAC promoter for the overproduction of proteins, recombinant clones were grown overnight in LB at 37°C in duplicate in three different induction conditions (IPTG concentrations): 0 mmol.L-1, 1 mmol.L-1 and 5 mmol.L-1. OD600 of each culture was measured every hour over six hours. E. coli NM522 strain was grown overnight in LB at 37°C in the same three induction conditions as control.Results
- Noise of the pTAC: fluorescence in absence of IPTG
We studied the noise of the promoter by comparing the normalized fluorescence between the construction and the NM522 strain without any induction of IPTG.

An ANOVA was made to see if there was a time effect between the two populations. We obtained a p-value of 0.61, suggesting that time had no effect on pTAC-RFP expression in absence of IPTG (α<0.05). Given this result, we gathered data to analyze if there was a significant difference between the two strains. A Student test was performed with the variance not equal. The p-value of 5.44*10-4 indicated that the strain carrying pTAC-RFP transcriptional fusion displayed a higher fluorescence than the control strain.
- pTAC induction by increasing concentration of IPTG
Then we studied the induction of the promoter by comparing the normalized fluorescence of the construction under the induction of [IPTG] = 0 and 1 mmol.L-1.

An ANOVA test was made to see if there was a time effect between the two populations. A p-value of 0.21 indicated that time had no effect on the fluorescence induction (α<0.05). Data was therefore gathered in order to compare the strain fluorescence with 1 mmol.L-1 IPTG and no IPTG. We realized a Student test with the variance not equal and obtained a p-value of 8.57*10-3, showing a difference of fluorescence due to the presence of IPTG in the medium.
Finally, we compared the expression of the pTAC-RFP transcriptional fusion in 2 concentrations of IPTG: 1 and 5 mmol.L-1.

An ANOVA was made to see if there was a time effect between the two populations. A p-value of 0.06 indicated that time had no effect (α<0.05). From there, we could gather the data to analyze if there was a significant difference between the two concentrations of IPTG.
We realized a Student test with the variance not equal and obtained a p-value of 0.61, indicating that no significant difference of fluorescence was observed with the rise of IPTG concentration.
Conclusion
In the case of RFP, the pTAC promoter seemed to not enable a gene tune because statistics showed that there was a significant noise, even in the absence of IPTG.
2. pTAC_CFP characterization
Description
pTAC promoter was taken from part part BBa_K864400 . This promoter is a hybrid of two operons: the trp and lac operons.This promoter is inducible by IPTG and commonly used in Escherichia coli for overproduction of proteins. Escherichia coli NM522 strain that we used in our lab constitutively express the LacIq protein, a strong pTAC promoter repressor. However, in absence of IPTG, we observed a strong leakage when plating our BBa_K1934000 transformants. Therefore, we decided to put a CFP reporter ORF under control of the pTAC promoter to characterize the promoter-driven transcriptional noise.
Expression of the pTAC-CFP fusion in presence of increasing amount of the inductor IPTG
Experimental design
The CFP coding sequence (BBa_E2020) was placed in silico under the control of the pTAC promoter (BBa_K864400), a strong RBS (BBa_B0030) and a bidirectional terminator (BBa_B0011). IDT performed the DNA synthesis and delivered the part as gBlock. The construct was cloned by conventional ligation into pSB1C3 plasmid and then transformed into E. coli NM522 strain. In order to study the efficiency of the pTAC promoter for the overproduction of proteins, recombinant clones were grown overnight in LB at 37°C in duplicate in three different induction conditions (IPTG concentrations): 0 mmol.L-1, 1 mmol.L-1 and 5 mmol.L-1. OD600 of each culture was measured every hour over six hours. E. coli NM522 strain was grown overnight in LB at 37°C in the same three induction conditions as control.Results
- Noise of the pTAC: fluorescence in absence of IPTG
We studied the noise of the promoter by comparing the normalized fluorescence between the construction and the NM522 strain without any induction of IPTG.

An ANOVA was made to see if there was a time effect between the two populations. We obtain a p-value of 0.96, suggesting that time had no effect on pTAC-CFP expression in absence of IPTG (α<0.05). Given this result, we gathered data to analyze if there was a significant difference between the two strains. A Student test was performed with the variance not equal. The p-value of 0.10 indicated that the strain carrying pTAC-CFP transcriptional fusion didn’t display a significant fluorescence difference with the control strain.
- pTAC induction by increasing concentration of IPTG
Then we studied the induction of the promoter by comparing the normalized fluorescence of the construction under the induction of [IPTG] = 0 and 1 mmol.L-1.

An ANOVA test was made to see if there was a time effect between the two populations. A p-value of 0.05 indicated that time had no effect on the fluorescence induction (α<0.05). Data was therefore gathered in order to compare the strain fluorescence with 1 mmol.L-1 IPTG and no IPTG. We realized a Student test with the variance not equal and obtained a p-value of 4.64*10-10, showing a significant difference of fluorescence due to the presence of IPTG in the medium.
Finally, we compared the expression of the pTAC-CFP transcriptional fusion in 2 concentrations of IPTG: 1 and 5 mmol.L-1.

An ANOVA was made to see if there was a time effect between the two populations. A p-value of 0.55 indicated that time had no effect (α<0.05). From there, we gathered data to analyze if there was a significant difference between the two concentrations of IPTG. We realized a Student test with the variance not equal and obtained a p-value of 0.54, indicating that no significant difference of fluorescence was observed with the rise of IPTG concentration.
Conclusion
In the case of CFP, the pTAC promoter seemed to enable a gene tune because there wasn’t a differential gene expression in absence of IPTG, so a significant noise wasn’t measured.
Team JUN_China 2019: Fluorescent characterization of the Tac promoter
Tac promoter is a hybrid of two operons: the trp and lac operons and it is inducible by IPTG.In our project, we built a number of parts which also contains a large number of construct intermediates using the tac promoter. The tac promoter is critical in our entire project, so we used green and red fluorescent protein to characterize its effectiveness. At the same time, we explored the changes in expression intensity of green and red fluorescent protein under the control of tac promoter by IPTG inducing at different times.
Experiment design:
We ligated the eGFP and RFP gene into the loop vector pZM1 (Ptac) containing the inducible tac promoter respectively and transfered eGFP and RFP into our chassis microorganism: Corynebacterium glutamicum F343 to carry out fermentation experiment. We added 1mmol/L IPTG to the fermentation broth at 0h, 1h, and 2h respectively and measured the fluorescence intensity and cell growth at different times to investigate whether the inducing capability of tac promoter was different at different induction times.
During the detection process, the cells were washed using PBS and the cells were diluted by appropriate multiples according to the measurement range of the instrument. The growth state and fluorescence intensity of the cells were detected using 96-well plates. The green fluorescent protein has an excitation wavelength of 488 nm, an emission wavelength of 517 nm. The red fluorescent protein excitation wavelength is 560 nm, and the emission wavelength is 630 nm.
Result:
We used the infinite 200Pro instrument to measure green and red fluorescent protein and OD600 measurement. The relevant data are shown below:
The data showed:
1.When inducing at 0 hour, the highest fluorescence value were at 16 hours, and there was a gradual decrease in data from 24 hours to 48 hours. This may due to the degradation of fluorescent protein.
2.The fluorescence values induced at 1 hour were relatively stable, but when at 16 hours the fluorescence value was best.
3.The expression of GFP was the worst when inducing at 2 hour.
Red fluorescent protein data showed that the expression of the red fluorescent protein was best when inducing at two hours and the value reached about 17,000 ua at 24 hours. The optimal induction time of red fluorescent protein expression was the same as the optimal induction time of polymerase in our project, which was induced at two hours after fermentation, and the product expression was highest. The data indicated that induction at different times, the expression of the target gene and the yield of the product were different.
Team GreatBay_SZ 2020
We have used pTac to express the key protein of our project: Geobacter sulfurreducens' pilin-based electrically conductive protein nanowires (e-PNs). The gene circuit includes this tac promoter which promotes the pilin gene(pilA) to express protein monomers; the monomers are later assembled into protein nanowires by the TypeIV pilin assembly system, which is constructed from the gene cluster at the downstream of the pilin gene(figure 1).

Figure 1: A Construction of BIOT gene circuit
The e-PN can be successfully expressed by express this part under the control of this pTac, and the result was verified by Western Blot(figure 2).