Difference between revisions of "Part:BBa K1806004"
| (2 intermediate revisions by one other user not shown) | |||
| Line 6: | Line 6: | ||
| − | Heat-shock is a major process for the survival of all species. In an environment where the temperature shows even the slightest inconsistency forces the organism to adapt to temperature shifts. This is where heat-shock proteins come into effect. Through sensing temperature shifts in the environment, organisms are able to survive and thrive in forever changing temperature conditions. | + | Heat-shock is a major process for the survival of all species.[1] In an environment where the temperature shows even the slightest inconsistency forces the organism to adapt to temperature shifts. This is where heat-shock proteins come into effect. Through sensing temperature shifts in the environment, organisms are able to survive and thrive in forever changing temperature conditions. |
| − | It has been understood that heat-shock mechanisms are required in almost all organisms and is present in the most primitive bacteria species to the most complex organisms. This very rudimentary mechanism therefore necessitates to be handled by the most common structures found in all that is living, RNA. The heat sensitive mechanisms in living organisms are conducted by | + | It has been understood that heat-shock mechanisms are required in almost all organisms and is present in the most primitive bacteria species to the most complex organisms.[2] This very rudimentary mechanism therefore necessitates to be handled by the most common structures found in all that is living, RNA. The heat sensitive mechanisms in living organisms are conducted by |
| − | + | ||
| − | |||
| + | RNA thermometers. | ||
| + | |||
| + | The Properties of RNA Thermometers | ||
RNA Thermometers are a group of RNA structures that have varying properties and operative functions. Although RNA Thermometers are basic and found in almost all organisms, characteristic differences exist in between RNA Thermometers. Between species, conformational, structural, functional and mechanical differences are present. This diversity allows the presence of many RNA Thermometer structures for application in different circumstances. | RNA Thermometers are a group of RNA structures that have varying properties and operative functions. Although RNA Thermometers are basic and found in almost all organisms, characteristic differences exist in between RNA Thermometers. Between species, conformational, structural, functional and mechanical differences are present. This diversity allows the presence of many RNA Thermometer structures for application in different circumstances. | ||
| − | There are common features of RNA Thermometers. The main feature of all RNA thermometers is that they function through conformational shifts in structure. These shifts cause conformational changes to expose the Shine-Dalgarno sequence, which acts as a binding site to allow translation. For translation to occur, the ribosome has to the aforementioned SD sequence. The structural differences are caused by the transcription regions, but the SD sequence is common. Aside from that, temperature is the factor responsible for changes in all RNA thermometers, but there are cold-shock RNA Thermometers as well as heat-shock. | + | There are common features of RNA Thermometers. The main feature of all RNA thermometers is that they function through conformational shifts in structure. These shifts cause conformational changes to expose the Shine-Dalgarno sequence, which acts as a binding site to allow translation.[3] For translation to occur, the ribosome has to the aforementioned SD sequence. The structural differences are caused by the transcription regions, but the SD sequence is common. Aside from that, temperature is the factor responsible for changes in all RNA thermometers, but there are cold-shock RNA Thermometers as well as heat-shock.[4] |
| + | |||
ibPB RNA Thermometer | ibPB RNA Thermometer | ||
| − | The Rna thermometer that will be utilized in our project is the ibpb RNA thermometer. Taken from the strains of the x species of the x group of organisms, the ibpb thermometer is a standard cis-acting heat-shock regulated rna thermometer. The heat shock process is stimulated by the medium of the attached protein. This heat shock response is effectively stimulated at 37 °C and beyond, however the RNA thermometer starts to function at 32 °C. 32 °C presents enough energy for some of the strands in the medium to start translation, while mostly, translation occurs at low levels. The unbinding of the dna sequence reaches its maximum at 37 °C, but the rate of the reaction and translation increases as the temperature further increases, as the number of ribosomes that can collusively attach to the SD sequence increase with temperature. This means that the activity of the strain increases with temperature, with the temperature of denaturation being the limiting factor. | + | The Rna thermometer that will be utilized in our project is the ibpb RNA thermometer. Taken from the strains of the x species of the x group of organisms, the ibpb thermometer is a standard cis-acting heat-shock regulated rna thermometer. The heat shock process is stimulated by the medium of the attached protein. This heat shock response is effectively stimulated at 37 °C and beyond, however the RNA thermometer starts to function at 32 °C.[5] 32 °C presents enough energy for some of the strands in the medium to start translation, while mostly, translation occurs at low levels. The unbinding of the dna sequence reaches its maximum at 37 °C, but the rate of the reaction and translation increases as the temperature further increases, as the number of ribosomes that can collusively attach to the SD sequence increase with temperature. This means that the activity of the strain increases with temperature, with the temperature of denaturation being the limiting factor. |
| Line 30: | Line 32: | ||
[[File:AUC_TURKEY_6_colony.png]] [[File:6004-2.png]] | [[File:AUC_TURKEY_6_colony.png]] [[File:6004-2.png]] | ||
| − | [[File:AUC_TURKEY_6,1, | + | [[File:AUC_TURKEY_6,1,3cutcheck.png]] [[File:6004-4.png]] |
== Functional Assay == | == Functional Assay == | ||
| Line 120: | Line 122: | ||
| − | < | + | |
| − | ===Functional Parameters== | + | |
| + | |||
| + | |||
| + | |||
| + | == References == | ||
| + | |||
| + | <html> | ||
| + | <font size="-10" face="arial"> | ||
| + | |||
| + | <p><font size="2"><b>1</b></font> Guisbert, E., Yura, T., Rhodius, V.A., and Gross, C.A. “Convergence of molecular, modeling and systems approaches for an understanding of the Escherichia coli heat shock response.” Microbiol. Mol. Biol. Rev. 72 (2008): 545-554. doi:10.1128/MMBR.00007-08 </p> | ||
| + | |||
| + | <p><font size="2"><b>2</b></font> Tripathy, Sindhu Nandini. "Molecular Biology of Endosalpinx." In The Fallopian Tubes, 31. New Delhi: Jaypee Brothers Medical Brothers, 2013.</p> | ||
| + | |||
| + | <p><font size="2"><b>3</b></font> Narberhaus F, Waldminghaus T, Chowdhury S. "RNA thermometers". FEMS Microbiol. Rev. 30 no.1 (2006):3–16. doi:10.1111/j.1574-6976.2005.004.x. PMID 16438677.</p> | ||
| + | |||
| + | <p><font size="2"><b>4</b></font> Jens Kortmann, Franz Narberhaus. “Bacterial RNA thermometers: molecular zippers and switches” Nature Reviews Microbiology 10 (2012):255-265</p> | ||
| + | |||
| + | <p><font size="2"><b>5</b></font> Shearstone, Jeffrey R., and Francois Baneyx. "Biochemical Characterization of the Small Heat Shock Protein IbpB from Escherichia Coli." THE JOURNAL OF BIOLOGICAL CHEMISTRY 274, no. 15 (1999): 9937-945.</p> | ||
| + | |||
| + | </font> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | |||
| + | ==Functional Parameters: Austin_UTexas== | ||
| + | <html> | ||
| + | <body> | ||
<partinfo>BBa_K1806004 parameters</partinfo> | <partinfo>BBa_K1806004 parameters</partinfo> | ||
| − | < | + | <h3><center>Burden Imposed by this Part:</center></h3> |
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/f/fa/T--Austin_Utexas--no_burden_icon.png" style = "width:160px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: 3.5 ± 4.5% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </body> | ||
| + | </html> | ||
Latest revision as of 19:16, 3 September 2020
T7+ ibPB RNA Thermometer + pelB + MCS + 6xHis
The part is to act as a backbone for the T7+ibPB RNA Thermometer+pelB+6xHis system. The MCS(Multiple Cloning Site) as the ligation region for the parts to be integrated into the system. The part that is added to the backbone will be translated in the presence of higher temperatures. The pelB signal peptide is present for the produced protein to be carried out of the cell. The 6xHis is for protein analysis.
Heat-shock is a major process for the survival of all species.[1] In an environment where the temperature shows even the slightest inconsistency forces the organism to adapt to temperature shifts. This is where heat-shock proteins come into effect. Through sensing temperature shifts in the environment, organisms are able to survive and thrive in forever changing temperature conditions.
It has been understood that heat-shock mechanisms are required in almost all organisms and is present in the most primitive bacteria species to the most complex organisms.[2] This very rudimentary mechanism therefore necessitates to be handled by the most common structures found in all that is living, RNA. The heat sensitive mechanisms in living organisms are conducted by
RNA thermometers.
The Properties of RNA Thermometers RNA Thermometers are a group of RNA structures that have varying properties and operative functions. Although RNA Thermometers are basic and found in almost all organisms, characteristic differences exist in between RNA Thermometers. Between species, conformational, structural, functional and mechanical differences are present. This diversity allows the presence of many RNA Thermometer structures for application in different circumstances.
There are common features of RNA Thermometers. The main feature of all RNA thermometers is that they function through conformational shifts in structure. These shifts cause conformational changes to expose the Shine-Dalgarno sequence, which acts as a binding site to allow translation.[3] For translation to occur, the ribosome has to the aforementioned SD sequence. The structural differences are caused by the transcription regions, but the SD sequence is common. Aside from that, temperature is the factor responsible for changes in all RNA thermometers, but there are cold-shock RNA Thermometers as well as heat-shock.[4]
ibPB RNA Thermometer
The Rna thermometer that will be utilized in our project is the ibpb RNA thermometer. Taken from the strains of the x species of the x group of organisms, the ibpb thermometer is a standard cis-acting heat-shock regulated rna thermometer. The heat shock process is stimulated by the medium of the attached protein. This heat shock response is effectively stimulated at 37 °C and beyond, however the RNA thermometer starts to function at 32 °C.[5] 32 °C presents enough energy for some of the strands in the medium to start translation, while mostly, translation occurs at low levels. The unbinding of the dna sequence reaches its maximum at 37 °C, but the rate of the reaction and translation increases as the temperature further increases, as the number of ribosomes that can collusively attach to the SD sequence increase with temperature. This means that the activity of the strain increases with temperature, with the temperature of denaturation being the limiting factor.
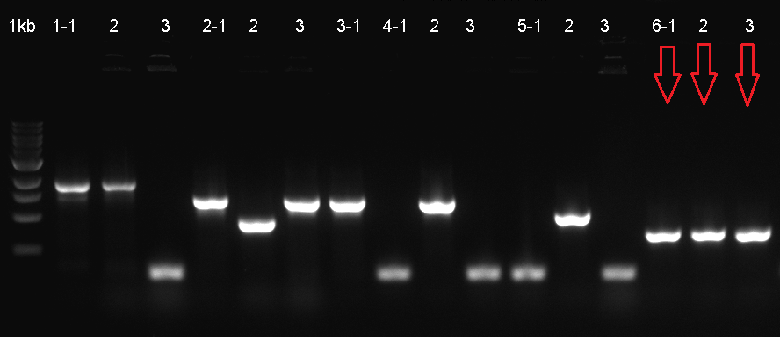
Cloning
The composite system was ligated to the cloning vector psB1C3, to be transformated and then verified with Colony PCR. The verified ligation was then moved on to the digestion step to be cut with the EcoR1 and Pst1 restriction enzymes. The gel run of the cut parts were portrayed down below. The part was tagged as G-Block 6 and sample 6-2 yielded accurate results.
Functional Assay
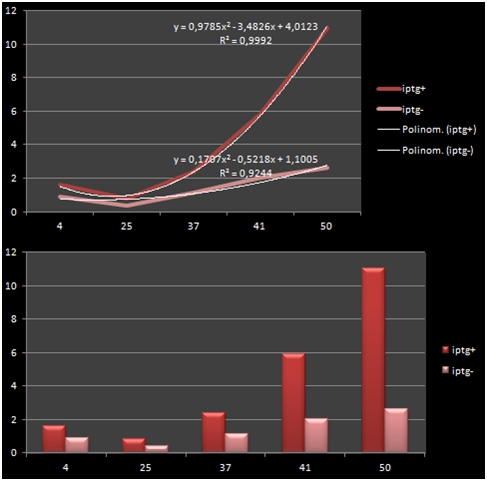
The bacteria that were transformated with G-Block 7 were cultured in 5 ml and incubated for 13 hours. After this incubation, the cultures were incubated at respective temperatures for 4 hours and were given 50uM of iPTG twice with 4 hours periouds. There were a total 5 different temperatures that the cultures were incubated in: 4, 25, 37, 42 and 50 C. The proteins in the cultures were then isolated and the protein concentrations were measured. Data on RFP concentration were acquired at 584 nm of emission and 607 nm of excitation. The acquired values were divided to the total amount of protein to acquire the following ratio.
| Medium Temperature | iPTG Presence +/- | Total Amount of Protein | RFP Fluorometric Measurement | RFP/Total |
|---|---|---|---|---|
| 4 | + | 14.09 | 22.32 | 1.584 |
| 4 | - | 11.349 | 10.36 | 0.912 |
| 25 | + | 17.87 | 14.35 | 0.802 |
| 25 | - | 11.054 | 4,313 | 0,390 |
| 37 | + | 16.808 | 40.2 | 2.391 |
| 37 | - | 15.216 | 17.36 | 1.140 |
| 41 | + | 7.637 | 44.83 | 5.870 |
| 41 | - | 5.557 | 11.13 | 2.002 |
| 50 | + | 5.463 | 60.06 | 10.993 |
| 50 | - | 7.997 | 20.95 | 2.619 |
The graph shows the variance in density concentrations of RFP, caused as a result of the functioning of the iPTG Inducible Promoters in different temperatures. The functioning of the promoters increased cumulatively with increased temperatures.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 2
Illegal BamHI site found at 252
Illegal XhoI site found at 277 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 219
- 1000COMPATIBLE WITH RFC[1000]
References
1 Guisbert, E., Yura, T., Rhodius, V.A., and Gross, C.A. “Convergence of molecular, modeling and systems approaches for an understanding of the Escherichia coli heat shock response.” Microbiol. Mol. Biol. Rev. 72 (2008): 545-554. doi:10.1128/MMBR.00007-08 2 Tripathy, Sindhu Nandini. "Molecular Biology of Endosalpinx." In The Fallopian Tubes, 31. New Delhi: Jaypee Brothers Medical Brothers, 2013. 3 Narberhaus F, Waldminghaus T, Chowdhury S. "RNA thermometers". FEMS Microbiol. Rev. 30 no.1 (2006):3–16. doi:10.1111/j.1574-6976.2005.004.x. PMID 16438677. 4 Jens Kortmann, Franz Narberhaus. “Bacterial RNA thermometers: molecular zippers and switches” Nature Reviews Microbiology 10 (2012):255-265 5 Shearstone, Jeffrey R., and Francois Baneyx. "Biochemical Characterization of the Small Heat Shock Protein IbpB from Escherichia Coli." THE JOURNAL OF BIOLOGICAL CHEMISTRY 274, no. 15 (1999): 9937-945.
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.