Difference between revisions of "Part:BBa K1758102"
m (→Characterization in vivo) |
|||
| (7 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1758102 short</partinfo> | <partinfo>BBa_K1758102 short</partinfo> | ||
| − | This part is based on [https://parts.igem.org/Part:BBa_I746909 BBa_I746909] but contains an optimized untranslated region (UTR), which improves the sfGFP expression <i>in vivo</i> as well as <i>in vitro</i>. This 5'-UTR, which improves the binding of the mRNA to the ribosome ([http://www.ncbi.nlm.nih.gov/pubmed/2676996 Olins et al. 1989]), is especially helpful in ''in vitro'' cell free protein synthesis. The 5'-UTR sequence contains a spacer of 10 adenine bases which has been shown to further enhance translation efficiency ([http://www.ncbi.nlm.nih.gov/pubmed/23927491 Takahashi et al. 2013]). The part also contains a double terminator made of [https://parts.igem.org/wiki/index.php?title=Part:BBa_B0010 B0010] and [https://parts.igem.org/wiki/index.php?title=Part:BBa_B0012 B0012]. | + | This part is based on [https://parts.igem.org/Part:BBa_I746909 BBa_I746909] but contains an optimized untranslated region (UTR, [https://parts.igem.org/Part:BBa_K1758100 BBa_K1758100]), which improves the sfGFP expression <i>in vivo</i> as well as <i>in vitro</i>. This 5'-UTR, which improves the binding of the mRNA to the ribosome ([http://www.ncbi.nlm.nih.gov/pubmed/2676996 Olins et al. 1989]), is especially helpful in ''in vitro'' cell free protein synthesis. The 5'-UTR sequence contains a spacer of 10 adenine bases which has been shown to further enhance translation efficiency ([http://www.ncbi.nlm.nih.gov/pubmed/23927491 Takahashi et al. 2013]). The part also contains a double terminator made of [https://parts.igem.org/wiki/index.php?title=Part:BBa_B0010 B0010] and [https://parts.igem.org/wiki/index.php?title=Part:BBa_B0012 B0012]. |
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | This part has been used in <i>E. coli</i>. When T7 polymerase is present, a fast production of sfGFP is observed. This part acts as a reliable reporter protein generator. | |
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
| Line 15: | Line 15: | ||
== Performance == | == Performance == | ||
<html> | <html> | ||
| − | <p> In our project, we developed cell-free biosensors with the help of <i>in vitro</i> protein synthesis. This part became our positive control due to </p> | + | <p> In our project, we developed cell-free biosensors with the help of <i>in vitro</i> protein synthesis. This part became our positive control due to superior performance compared to <a href="https://parts.igem.org/Part:BBa_I746909">I746909</a> </p> |
| − | <p> We were sure that a further optimization of sfGFP production was possible. Based on literature screening (for details see <a href="http://2015.igem.org/Team:Bielefeld-CeBiTec/Project/CFPS">background page</a>, we designed a <b>translation enhancing sequence (5'-untranslated region, 5'-UTR)</b> and inserted it into P<sub>T7</sub>-sfGFP, thereby improving it. The insertion led to the creation of this part. </p> | + | <p> We were sure that a further optimization of sfGFP production was possible. Based on literature screening (for details see <a href="http://2015.igem.org/Team:Bielefeld-CeBiTec/Project/CFPS">background page</a>), we designed a <b>translation enhancing sequence (5'-untranslated region, 5'-UTR, BBa_K1758100)</b> and inserted it into P<sub>T7</sub>-sfGFP, thereby improving it. The insertion led to the creation of this part. </p> |
<p> We designed the 5'-UTR on assumption that if translation was a bottleneck in our cell-free protein synthesis system, this sequence would improve sfGFP production. As verification for the beneficial effect of the 5'-UTR, we performed <i>in vivo</i> experiments, normalizing the fluorescence signal to culture OD<sub>600</sub>. | <p> We designed the 5'-UTR on assumption that if translation was a bottleneck in our cell-free protein synthesis system, this sequence would improve sfGFP production. As verification for the beneficial effect of the 5'-UTR, we performed <i>in vivo</i> experiments, normalizing the fluorescence signal to culture OD<sub>600</sub>. | ||
| Line 23: | Line 23: | ||
</html> | </html> | ||
| − | |||
==Characterization <i>in vivo</i>== | ==Characterization <i>in vivo</i>== | ||
| − | [[File:Bielefeld-CeBiTec_CFPS_UTR.png|thumb|600px|center|In vivo characterization of sfGFP with and | + | [[File:Bielefeld-CeBiTec_CFPS_UTR.png|thumb|600px|center|In vivo characterization of sfGFP with and without our designed, translation enhancing 5'-untranslated region (5'-UTR; BBa_K1758100). Relative fluorescence units were normalized on OD600. Error bars represent standard deviation of triplicates. PT7-sfGFP: BBa_I746909; PT7-UTR-sfGFP: BBa_K1758102 (This part)]] |
| Line 34: | Line 33: | ||
| − | [[File:Bielefeld-CeBiTec_CFPS_UTR_cultures.png|thumb|400px|center|Comparision of cultures expressing sfGFP with the optimized UTR and without. Left: BBa_K1758102, right: BBa_I746909]] | + | [[File:Bielefeld-CeBiTec_CFPS_UTR_cultures.png|thumb|400px|center|Comparision of cultures expressing sfGFP with the optimized UTR and without. Left: BBa_K1758102 (This part), right: BBa_I746909]] |
| − | + | ||
==Characterization <i>in vitro</i>== | ==Characterization <i>in vitro</i>== | ||
[[File:Bielefeld-CeBiTec_CFPS_UTR_invitro.png|thumb|400px|center|Importance of 5'-untranslated region (UTR) for in vitro protein synthesis. T7 refers to T7 promoter. sfGFP lysate refers to cell lysate won by sonication; cells were induced to produce sfGFP in vivo. Decreasing sfGFP lysate signal probably relied on evaporation. Mean values from two technical replicates are shown]] | [[File:Bielefeld-CeBiTec_CFPS_UTR_invitro.png|thumb|400px|center|Importance of 5'-untranslated region (UTR) for in vitro protein synthesis. T7 refers to T7 promoter. sfGFP lysate refers to cell lysate won by sonication; cells were induced to produce sfGFP in vivo. Decreasing sfGFP lysate signal probably relied on evaporation. Mean values from two technical replicates are shown]] | ||
| + | |||
| + | |||
| + | <html> | ||
| + | |||
| + | <p>As can be seen in the figure below, we improved <a href="https://parts.igem.org/Part:BBa_I746909">BBa_I746909</a> (PT7-sfGFP in the figure) by employing of the translation enhancing 5'-UTR (PT7-UTR-sfGFP in the figure) we designed. High yields for <i>in vitro</i> sfGFP production were only observed when 5'-UTR was present. </p> | ||
| + | |||
| + | |||
| + | <figure> | ||
| + | <a href="https://static.igem.org/mediawiki/2015/7/7e/Bielefeld-CeBiTec_150729_CFPS_T7-UTR-sfGFP.png" data-lightbox="group_150729" data-title="CFPS kinetics of sfGFP in cell extract"><img src="https://static.igem.org/mediawiki/2015/7/7e/Bielefeld-CeBiTec_150729_CFPS_T7-UTR-sfGFP.png" target="_blank" alt="CFPS in Tecan plate reader"></a> | ||
| + | <figcaption> Tracing CFPS kinetics in Tecan platereader. RFU signals were normalized to first signal of sfGFP lysate. Purified GFP refers to the signal of a His-tagged GFP solution at a concentration of about 320 µg/mL we kindly received from our advisor Lukas. Error bars represent standard deviation of triplicates. </figcaption> | ||
| + | </figure> | ||
| + | |||
| + | </html> | ||
| + | |||
| + | |||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
<partinfo>BBa_K1758102 parameters</partinfo> | <partinfo>BBa_K1758102 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
Latest revision as of 19:16, 19 September 2015
Translation enhancing 5-UTR + sfGFP under control of T7 promoter
This part is based on BBa_I746909 but contains an optimized untranslated region (UTR, BBa_K1758100), which improves the sfGFP expression in vivo as well as in vitro. This 5'-UTR, which improves the binding of the mRNA to the ribosome ([http://www.ncbi.nlm.nih.gov/pubmed/2676996 Olins et al. 1989]), is especially helpful in in vitro cell free protein synthesis. The 5'-UTR sequence contains a spacer of 10 adenine bases which has been shown to further enhance translation efficiency ([http://www.ncbi.nlm.nih.gov/pubmed/23927491 Takahashi et al. 2013]). The part also contains a double terminator made of B0010 and B0012.
Usage and Biology
This part has been used in E. coli. When T7 polymerase is present, a fast production of sfGFP is observed. This part acts as a reliable reporter protein generator.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 82
Performance
In our project, we developed cell-free biosensors with the help of in vitro protein synthesis. This part became our positive control due to superior performance compared to I746909
We were sure that a further optimization of sfGFP production was possible. Based on literature screening (for details see background page), we designed a translation enhancing sequence (5'-untranslated region, 5'-UTR, BBa_K1758100) and inserted it into PT7-sfGFP, thereby improving it. The insertion led to the creation of this part.
We designed the 5'-UTR on assumption that if translation was a bottleneck in our cell-free protein synthesis system, this sequence would improve sfGFP production. As verification for the beneficial effect of the 5'-UTR, we performed in vivo experiments, normalizing the fluorescence signal to culture OD600.
Characterization in vivo
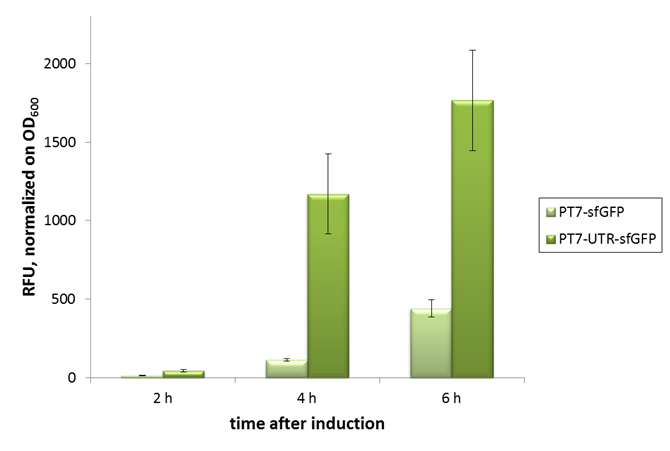
As can be seen in the picture, the difference was observable with the naked eye as well.
Characterization in vitro

As can be seen in the figure below, we improved BBa_I746909 (PT7-sfGFP in the figure) by employing of the translation enhancing 5'-UTR (PT7-UTR-sfGFP in the figure) we designed. High yields for in vitro sfGFP production were only observed when 5'-UTR was present.


