Difference between revisions of "Help:Protocols/3A Assembly"
m |
m (→Obtaining materials) |
||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | [[Category:BioBrick RFC 10]][[Category:Protocols]] | ||
{{HelpPage/MainLinks}} | {{HelpPage/MainLinks}} | ||
{{HelpPage/MainCSS}} | {{HelpPage/MainCSS}} | ||
| Line 4: | Line 5: | ||
==Before You Start!== | ==Before You Start!== | ||
| + | [[Image:3AAssembly.png|thumb|right|300px|3A Assembly Overview]] | ||
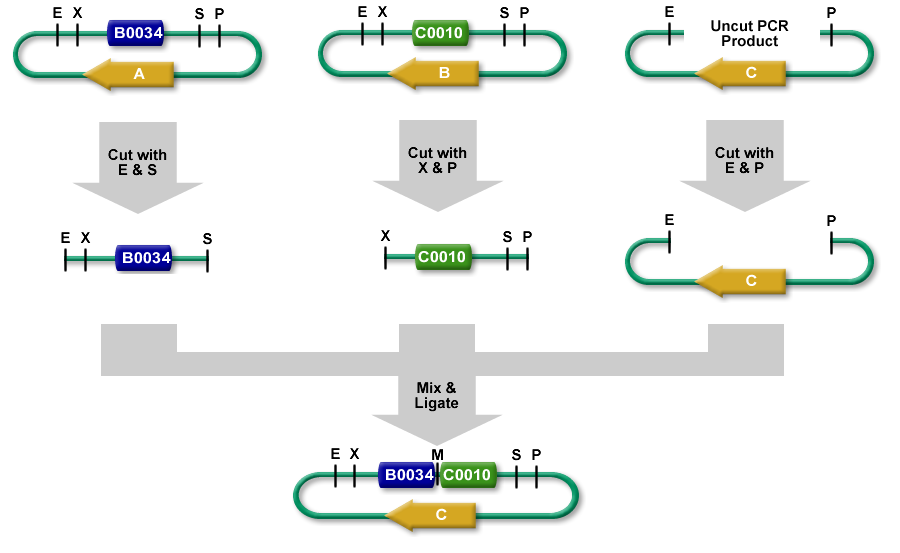
You should read the [[Help:Assembly/3A_Assembly|overview of 3A Assembly]] if you are not familiar with its mechanics. The protocol assumes that you have the part samples you will assemble ready in purified form (Miniprepped or PCR purified). The Protocol has has 4 stages: Restriction Digest, Ligation, Transformation, Picking Colonies. | You should read the [[Help:Assembly/3A_Assembly|overview of 3A Assembly]] if you are not familiar with its mechanics. The protocol assumes that you have the part samples you will assemble ready in purified form (Miniprepped or PCR purified). The Protocol has has 4 stages: Restriction Digest, Ligation, Transformation, Picking Colonies. | ||
| Line 31: | Line 33: | ||
** 5 ul NEB Buffer 2 | ** 5 ul NEB Buffer 2 | ||
** 0.5 ul BSA | ** 0.5 ul BSA | ||
| − | ** 0.5 ul | + | ** 0.5 ul EcoRI-HF |
| − | ** 0.5 ul | + | ** 0.5 ul PstI |
| − | ** 0.5 ul | + | ** 0.5 ul DpnI (Used to digest any template DNA from production) |
** 18 ul dH20 | ** 18 ul dH20 | ||
<html></td> | <html></td> | ||
| Line 82: | Line 84: | ||
* Add equimolar amount of '''Part A''' (EcoRI-HF SpeI digested) fragment (< 3 ul) | * Add equimolar amount of '''Part A''' (EcoRI-HF SpeI digested) fragment (< 3 ul) | ||
* Add equimolar amount of '''Part B''' (XbaI PstI digested fragment) (< 3 ul) | * Add equimolar amount of '''Part B''' (XbaI PstI digested fragment) (< 3 ul) | ||
| − | * Add 1 ul | + | * Add 1 ul T4 DNA ligase buffer. '''Note:''' Do not use quick ligase |
| − | * Add 0.5 ul | + | * Add 0.5 ul T4 DNA ligase |
* Add water to 10 ul | * Add water to 10 ul | ||
* Ligate 16C/30 min, heat kill 80C/20 min | * Ligate 16C/30 min, heat kill 80C/20 min | ||
| Line 94: | Line 96: | ||
===Obtaining materials=== | ===Obtaining materials=== | ||
| − | + | {{HelpPage/Updates/Requests}} | |
| − | + | ||
===Troubleshooting=== | ===Troubleshooting=== | ||
Latest revision as of 16:55, 31 May 2023
- Registry Help Pages:
- TOC
- At-a-Glance
- FAQ
Before You Start!
You should read the overview of 3A Assembly if you are not familiar with its mechanics. The protocol assumes that you have the part samples you will assemble ready in purified form (Miniprepped or PCR purified). The Protocol has has 4 stages: Restriction Digest, Ligation, Transformation, Picking Colonies.
Since you'll be assembling two part samples together, you will want to make sure you're putting them in the correct order. We will refer to the first part as Part A, and the second part Part B
Restriction Digest
Materials
- Your two Part Samples, A and B: Miniprepped DNA (in BioBrick RFC[10] plasmid backbones)
- Linearized Plasmid Backbone (with a different resistance to the plasmid backbones containing your part samples)
- EcoRI, XbaI, SpeI, PstI, DpnI
- NEB Buffer 2
- BSA
- dH20
Digest
|
|
|
|
|
|
|
|
||
Ligation
- Add 2ul of digested Plasmid Backbone (25 ng)
- Add equimolar amount of Part A (EcoRI-HF SpeI digested) fragment (< 3 ul)
- Add equimolar amount of Part B (XbaI PstI digested fragment) (< 3 ul)
- Add 1 ul T4 DNA ligase buffer. Note: Do not use quick ligase
- Add 0.5 ul T4 DNA ligase
- Add water to 10 ul
- Ligate 16C/30 min, heat kill 80C/20 min
- Transform with 1-2 ul of product
Note: For linearized plasmid backbones provided by iGEM HQ, a plasmid backbone with an insert of BBa_J04450 was used as template. As a result any red colonies that appear during your ligation may be due to the template as a background. Digesting with Dpn1 before use should reduce this occurrence.
Notes
Obtaining materials
iGEM HQ and the iGEM Registry no longer prepares and ships part and component requests to teams and labs. Information related to part and component requests on the Registry and other iGEM pages is either deprecated or kept for archival purposes.
We encourage our iGEM teams to use our sponsor synthesis offers to synthesize the samples you need, or to find them through the iGEM distribution kits, if available.
Troubleshooting
- In our experience, most problems with 3A Assembly come from the transformation process. Because 3A Assembly uses a triple ligation, the concentration of assembled DNA is about 10% of that produced by a double ligation process. Particularly, if your lab makes its own competent cells (and particularly chemically competent cells) it is important to test the competency. If you are having trouble obtaining colonies, we suggest that you try commercial competent cells. Be sure to follow the instructions carefully. Do not add more ligation product than specified and be careful when resuspending the competent cells.
- If you are interested in making your own competent cells, we suggest using the protocol that was developed by Tom Knight and used by the Registry.
- The second most common problems are caused by defects in the DNA entering the ligation. We find ligation to be a very reliable process. However, the sticky ends of the cut parts can be destroyed by enzymes. Also, the original DNA may be incorrect or defective.
- Another common mechanism for failure involves a piece of genomic DNA being cloned into the construction plasmid. This result typically occurs when the assembly is somehow unfavorable to the cell. This issue can be partially checked via the colony PCR step in the protocol though if the genomic DNA is of the same size as your assembly, then the single colony PCR screen is ineffective. A careful purification of the construction plasmid can help to avoid this result as can phosphatase treatment of the linear construction plasmid.
Analysis
- Since the transformed cells are plated on plates supplemented with the antibiotic corresponding to the construction plasmid, only cells containing the construction plasmid backbone should survive.
- Since the construction plasmid itself has been reduced in quantity by PCR and digestion by DamI, few colonies will contain the original construction plasmid.